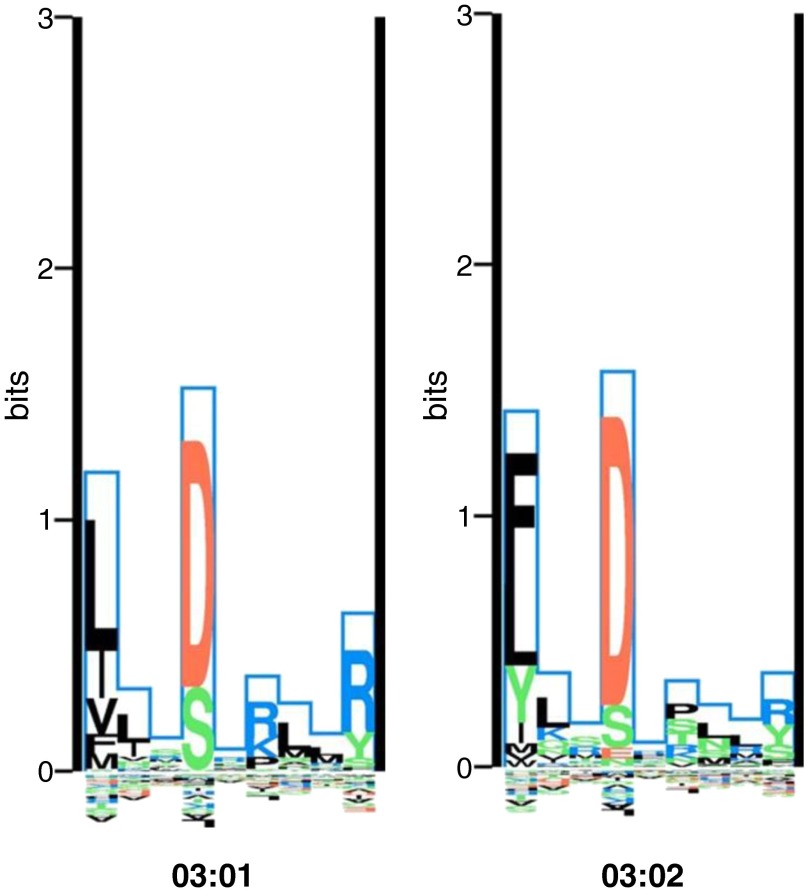
Figure 2.
The binding motifs of the HLA-DRB1*03:01 and 03:02 alleles visualized using a logo-plot. The Kullback–Leibler (KL) sequence logo representation of the binding motifs of the HLA-DRB1*03:01 and *03:02 alleles is taken from the MHC Motif Viewer website (http://www.cbs.dtu.dk/biotools/MHCMotifViewer/Home.html). The KL information content is plotted along the nine-amino-acid binding core. Amino acids with greater binding preference are plotted on the positive y axis, whereas amino acids with a negative influence on binding are plotted on the negative y axis. The height of each amino acid represents its relative binding specificity. Color-coding of amino acids represent the physicochemical properties of acidic (red), basic (blue), hydrophobic (black), and neutral (green). Of the primary anchor positions (P1, P4, P6, and P9), positions 1 and 4 show the most dramatic differences in amino acid preferences between the two HLA molecules. At P1, *03:01 prefers hydrophobic amino acids, whereas *03:02 prefers hydrophobic but will also bind neutral tyrosine. The allele *03:01 has a preference for basic amino acids at P6, whereas *03:02 is more promiscuous, preferentially binding with hydrophobic or neutral amino acids at the P6 anchor.