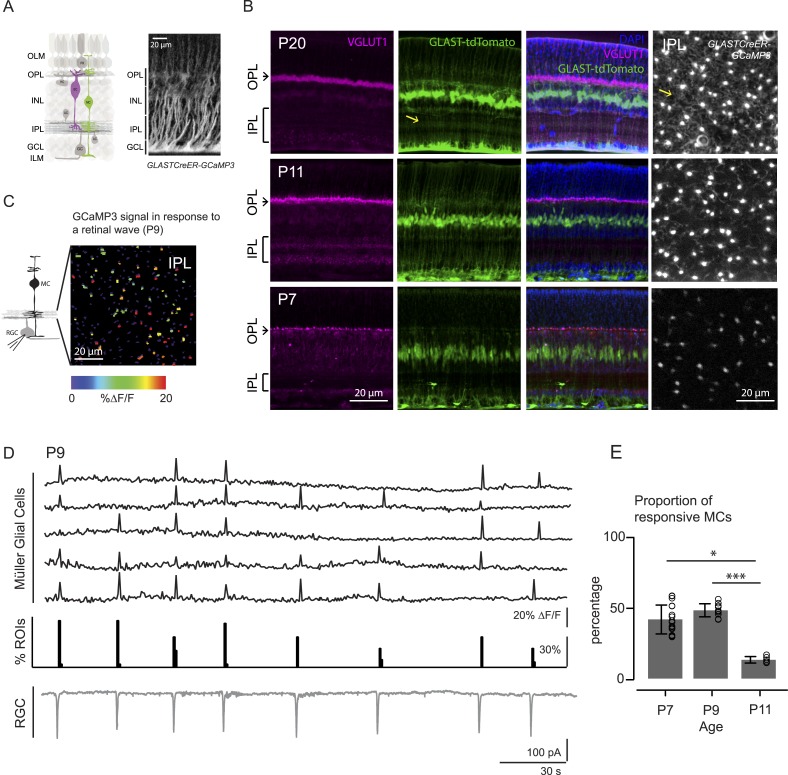
Figure 1. Morphology of Müller glial cells and their interactions with neurons change over development.
(A) Left, Diagram of adult retinal cross-section illustrates layered circuitry (OLM: outer limiting membrane; OPL: outer plexiform layer; INL: inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer; ILM: inner limiting membrane) and main cell types (PR: photoreceptor; HC: horizontal cell; BC: bipolar cell; MC: Müller glial cell; AC: amacrine cell; GC: ganglion cell). Right, Orthogonal projection of two-photon Z-stacks shows GCaMP3 expression in MCs of a P22 GLASTCreER::GCaMP3 mouse retina. (B) Vibratome sections of GLASTCreER::tdTomato retinas show the structure of MCs (green; tdTomato) and the expression of vesicular glutamate transporter 1 (VGLUT1) in bipolar cells (magenta; anti-VGLUT1) at different ages. Blue stain is 4′,6-diamidino-2-phenylindoele (DAPI) for visualizing cell nuclei as landmarks. Rightmost images are XY planes of the IPLs in GLASTCreER::GCaMP3 retinas showing GCaMP3 signal at different ages. Note the expansion of MC lateral processes into the IPL with development. Yellow arrows indicate lateral processes of the Müller glial cells. (C) Left, Circuit diagram of the retina highlights cells recorded for figures C and D; labeling as in Figure 1A. Right, XY plane shows GCaMP3 signals of MCs in response to a retinal wave in a P9 GLASTCreER::GCaMP3 retina. Color scale indicates normalized changes in fluorescence during a retinal wave. (D) Simultaneous MC calcium imaging and retinal ganglion cell (RGC) whole-cell voltage-clamp recording (Vm = −60 mV) of a P9 GLASTCreER::GCaMP3 retina. Sample ∆F/F traces (black traces) from individual regions of interests (ROIs) (that include stalks and processes of the MC population) in response to neuronal waves recorded in a RGC (grey trace). Histogram in middle denotes percentage over time of ROIs with responsive MCs. (E) Percentage of ROIs with responsive MCs during at least one retinal wave at different ages. P7: 1326 ROIs from 11 retinas; P9: 3027 ROIs from 14 retinas; P11: 872 ROIs from 6 retinas. Kruskal–Wallis one-way ANOVA, Dunn's post-hoc test. ***p < 0.001 and *p < 0.05. See also Figure 1—figure supplement 1 and Video 1.