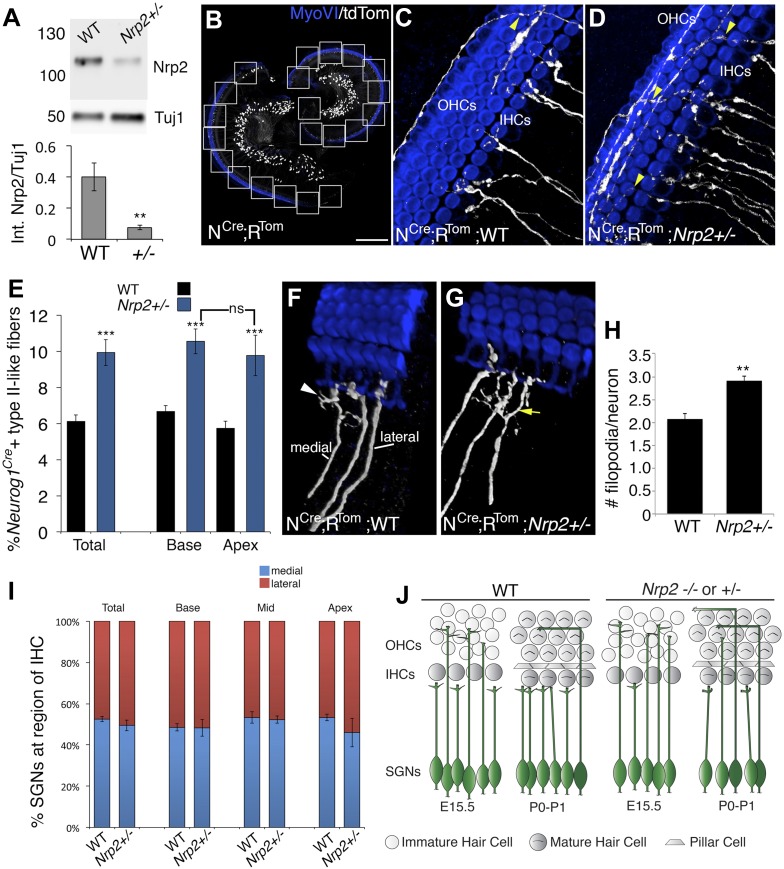
Figure 4. Nrp2 regulates SGN axonal complexity.
(A) Western blot demonstrating significantly reduced Nrp2 protein in cochleae from Nrp2+/− mice as compared to WT. **p ≤ 0.01; n = 9 WT; 7 Nrp2+/−. (B) Low-magnification view of a Neurog1CreERT2; R26R-tdTomato cochlea with boxed regions showing the locations where confocal z-stacks were acquired. In all of the micrographs in this figure, white represents anti-tdTomato and blue represents anti-myosin VI. (C, D) Compared to WT, Nrp2+/− cochleae with sparse SGN labeling show higher numbers of fibers in the OHC region (yellow arrowheads). (E) Quantification of the percentage of sparsely labeled type II-like fibers (defined as SGNs that had crossed into the OHC region and turned toward the base) in each genotype. ***p ≤ 0.001; n = 9 WT; 6 Nrp2+/−; ns, not significant. (F, G) 3D reconstructions of type I SGNs (relative positions are indicated in F). In Nrp2+/− cochleae, there were many type I SGNs with increased branching (yellow arrow in G), suggesting absence of an inhibitory cue. (H) Quantification of number of filopodia per individual type I SGN in WT and Nrp2+/− cochleae indicates significantly higher numbers of small branches in Nrp2+/− heterozygotes. **p ≤ 0.01; n = 4 WT; 6 Nrp2+/−. (I) The number of medially and laterally positioned type I SGNs does not change in Nrp2+/− cochleae. n = 4 cochleae each genotype. (J) Model illustrating phenotypic changes in cochlear innervation as a result of a decrease or absence of Nrp2. Error bars, sem. Scale bar in B: 150 μm, B; 20 μm, C, D, F, G.