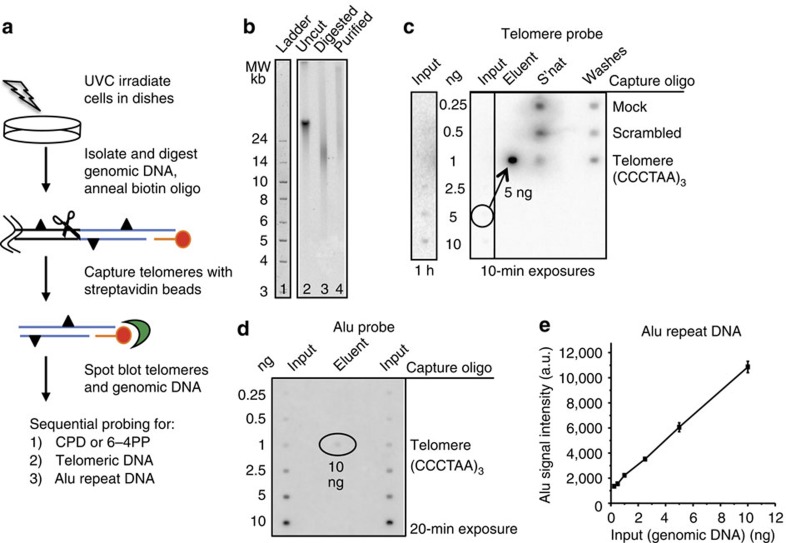
Figure 1. Telomere isolation assay.
(a) Schematic of telomere capture assay. Telomeres (blue lines) are released by digesting the genome (scissors) and are captured by annealing a biotinylated oligonucleotide (red) that binds to the telomeric single-strand overhang and to streptavidin beads (green). Triangles denote photoproducts. (b) Undigested (lane 2) and digested (lane 3) genomic DNA, and isolated telomeres (lane 4) from BJ-hTERT cells were electrophoresed on a 0.6% agarose gel that was subsequently hybridized with a radiolabelled telomeric probe (lane 2–4). The ladder was visualized by SYBR Green staining (lane 1). (c) Specificity of telomere capture. Telomeres were isolated using three different conditions: mock (no oligonucleotide), scrambled (non-telomeric oligonucleotide) and telomere oligonucleotide (Table 1). Various amounts of digested genomic DNA (input), 50% of the unbound (s'nat) and 50% of the combined washes were loaded on the membrane. 50% of the eluent for the telomere oligo (exactly 5 ng) and total eluent for the mock (0 ng) and scrambled oligo (2.6 ng) was loaded. The membrane was hybridized with a radiolabelled telomeric probe and exposed to a phosphoimager screen for one hour or 10 min as indicated. (d) Telomere purity. Various amounts of digested genomic DNA (input) and exactly 10 ng of the telomere eluent were loaded on a membrane that was hybridized with a radiolabelled Alu repeat DNA probe. (e) Alu signal intensities from the genomic DNA were plotted against the DNA amounts loaded. Values and error bars represent the mean and s.d. from two independent experiments. The Alu signal intensity for 10 ng of telomere eluent corresponded to about 1.2 ng.