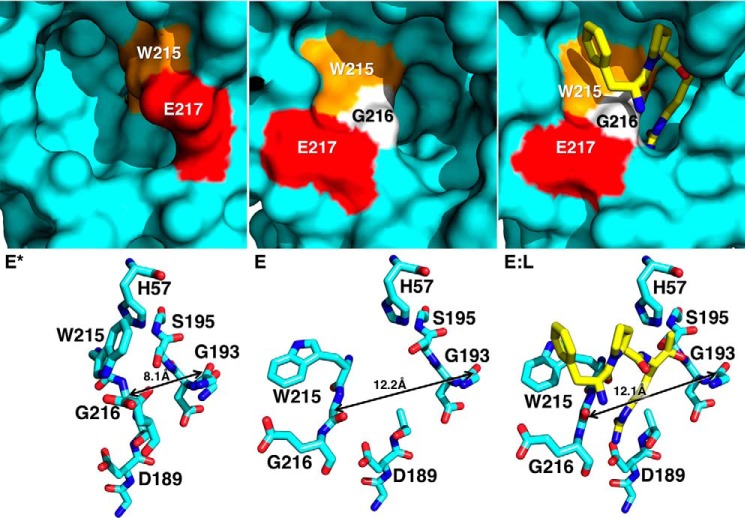
FIGURE 4.
X-ray crystal structures of thrombin mutant Y225P in the E* (Protein Data Bank 3S7H), E (Protein Data Bank 3S7K), and E:L (Protein Data Bank 1THP) conformations. Top panels depict surface representations of the active site of Y225P in three different conformations: free and closed (E*), free and open (E), and active site bound to PPACK (E:L). In the E* conformation the segment comprising residues Trp-215 (orange), Gly-216 (white), and Glu-217 (red) collapses into the active site, stabilized by interactions between His-57 and Trp-215 (bottom left). In the E conformation the same segment opens up, extending the aperture between the Cα atoms of residues Gly-193 and Gly-216 from 8.1 Å in the E* conformations to more than 12 Å in the E and EL conformations. The difference between the E* and E forms is significant, because PPACK (yellow) requires the wider 12-Å aperture to bind productively to the active site (bottom right).