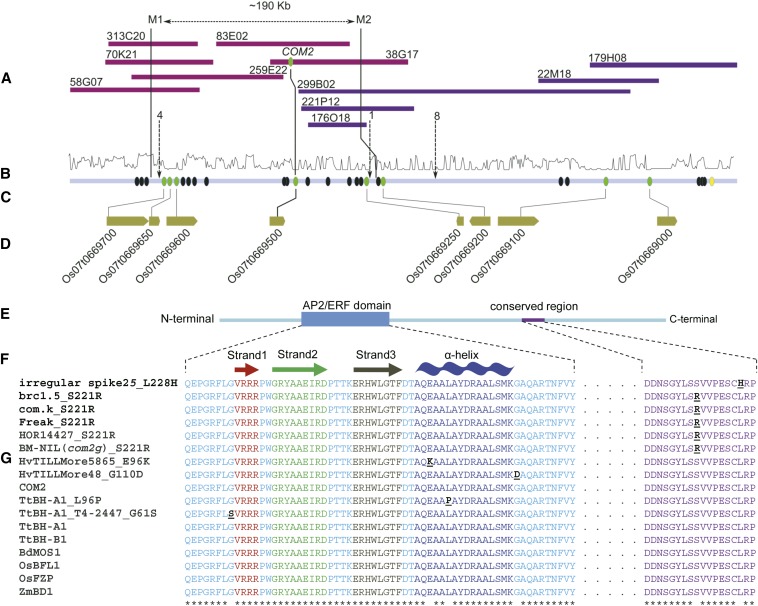
Figure 2.
High-resolution genetic linkage map of COM2 region on chromosome 2HS of barley and protein domain structure conservation among grasses. (A) Overlapped BAC clones (clones of the minimal tiling path) originated from two barley physical map contigs 44575 (purple) and 47813 (blue). The two contigs were merged as they showed significant edge sequence homology. (B) Depicts k-mer method-based repeat frequency (log-scaled; 0, 10). (C) Respective and predicted consensus sequence derived from BAC sequences in which circles represent either the Augustus gene model without sequence homology to Brachypodium, rice, sorghum, or barley genes (black), or with sequence homology to Brachypodium, rice, sorghum, or barley gene (green) as well as those with only sequence homology to predicted barley genes (yellow). The one-directional arrows connect the number of recombination to the corresponding position. (D) Rice genes syntenic to detected barley genes. (For corresponding low-resolution genetic mapping, see Figure S2 and File S2). (E) Protein sequence of COM2 with AP2/ERF DNA-binding domain and a highly conserved terminal region. (F) Position and structure of the AP2/ERF subdomains; including β-sheet (consists of strands 1, 2, and 3) and α-helix. (G) Alignment of AP2/ERF domain and conserved terminal region of COM2/ BHt-A1 with other grass orthologs and mutant alleles in barley and wheat. (F and G) Functional amino acid substitutions are underlined and shown in black. Asterisks indicate no amino acid changes at the corresponding position. For phenotype of respective mutant wheat and barley, see Figure S4.