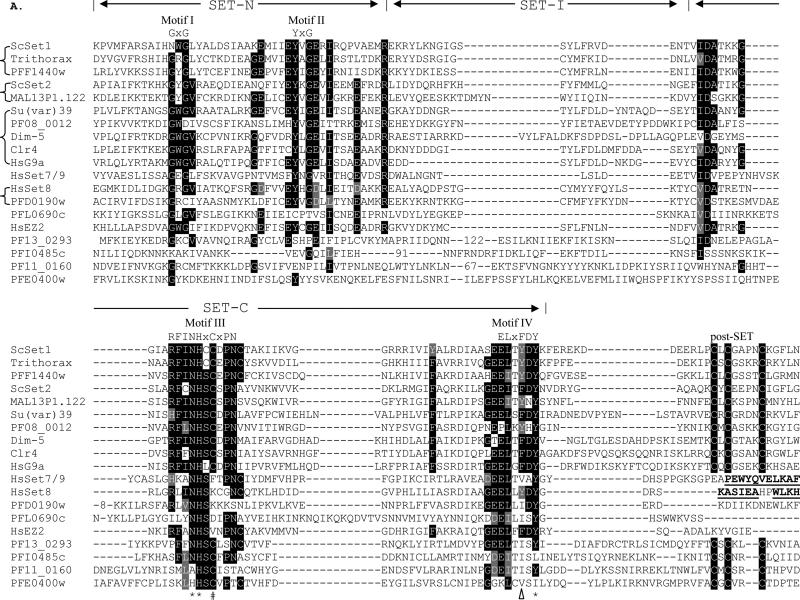
Fig. 2.

Sequence comparison of the SET and pre-SET domains. A) Alignment of SET domains of PfSETs with representatives of SET histone lysine methyltransferases. The three regions are indicated as SET-N, SET-I and SET-C. Signature motifs I-IV with the motif consensus sequences are indicated above the alignment. Numbers in the SET-I region indicate deleted residues for optimal alignments. Identical residues are shaded in black and conserved substitutions are shaded in grey. Three residues with catalytic roles are indicated with asterisks (*). The F/Y switch that results in product specificity of human SET7/9 and ScSET1 is indicated with a triangle. The Cys residue in motif III that is involved in the Zn cluster formation with the post-SET domain is indicated with a hash symbol (#). The C-terminal sequences that form an α-helical structure in human SET7/9 and SET8 are in bold and underlined. Representatives of different SET families from model organisms include ScSET1 (GenBank # EDN62358), ScSET2 (#P46995), Drosophila TRX (#CAA83515) and Su(var)39 (#NP_524357), Neurospora crassa Dim-5 (#AAL35215), Schizosaccharomyces pombe Clr4 (#CAA22283), HsG9a (#CAA49491), HsSET7/9 (#AAI21056), HsSET8 (#AAM47033), and HsEZ2 (#NP_004447). Abbreviations: Saccharomyces cerevisiae – Sc, Homo sapiens – Hs. B) Alignment of the pre-SET domain in PfSET3 and other members of the Su(var)39 subfamily with the Cys residues highlighted.