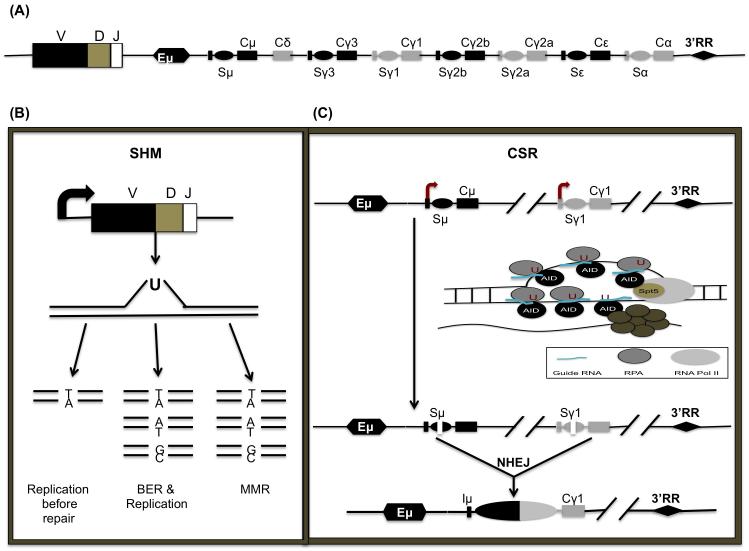
Figure 1.
Targeting of AID to the immunoglobulin locus (Ig): (A) Structure of the Ig heavy-chain locus after V(D)J recombination. The variable region V(D)J is followed by a series of constant regions, each specifying a different antibody isotype. Each constant region gene is comprised of a transcription unit with a cytokine-inducible promoter (P), an intervening (I)-exon, S region, and CH exons. The C regions are preceded by corresponding switch (S) repetitive sequences. Distal enhancers are present at the 3′ end of the IgH locus. (B) During SHM, mutations are introduced into the rearranged variable region genes. SHM begins with transcription through the variable region. ssDNA is created. If replication begins prior to the replacement of the U, a transition mutation (Ts) will appear at the original G:C to T:A. If BER removes the U prior to replication, Ts or Tv (transversion) mutations will replace the G:C. Finally, if the U is removed by the MMR pathway, both Ts and Tv mutations will occur at G:C and A:T base pairs. (C) CSR is initiated by transcription through the S regions, which leads to the formation of stable R-loops. RNA polII stalls at various regions due to the formation of secondary DNA structures like R loops. Following stalling, AID interacts with its cofactor, Spt5 and guide RNA’s which target AID to complementary S region DNA based on sequence information provided by the guide RNAs. The ssDNA is stabilized by the ssDNA binding protein complex RPA. Binding of the exosome degrades or displaces the nascent transcript to generate ssDNA, an AID substrate on the template strand.