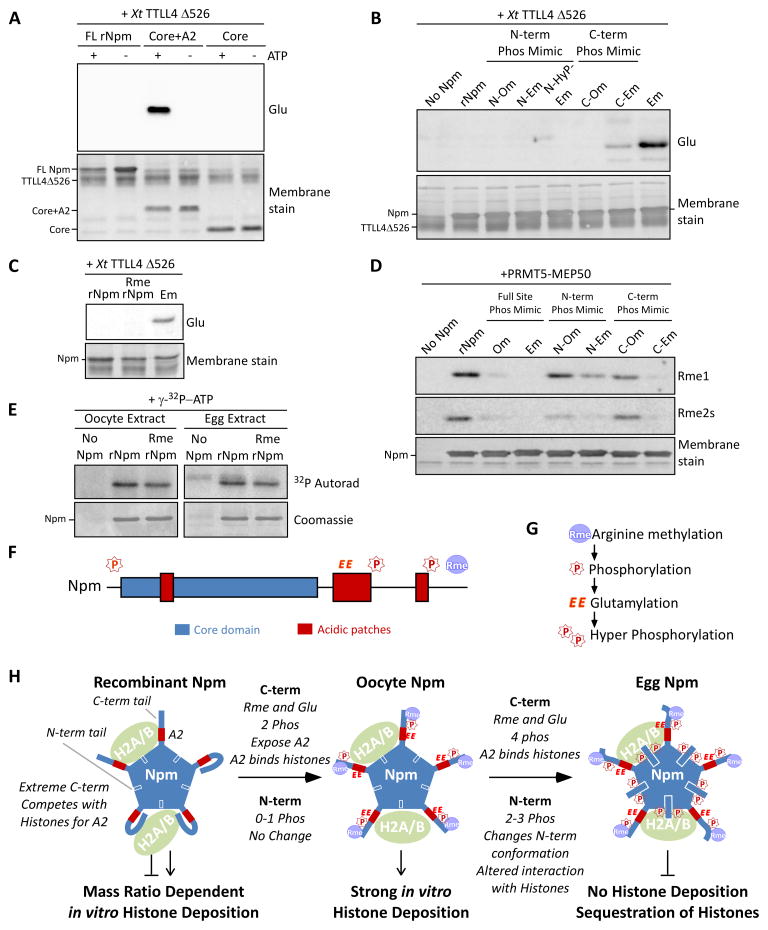
Figure 7. Nucleoplasmin phosphorylation, glutamylation, and methylation crosstalk reveals a developmental mechanism for regulated histone accessibility.
A. X. tropicalis TTLL4Δ526 glutamylation of Npm truncation mutants. Immunoblot for glutamylation (Glu) and Direct Blue 71 membrane stain for total protein are shown. B. Glutamylation assay using phosphomimetic mutants of Npm as in (A). C. Glutamylation assay using PRMT5-treated rNpm (“Rme rNpm”) and Em phosphomimetic mutant, as in (A). D. Arginine methylation assay using phosphomimetic mutants of Npm and XlPRMT5-MEP50. Immunoblot for mono- (Rme1) and symmetric dimethylarginine (Rme2s) and the membrane stain are shown. E. Kinase assay of rNpm and arginine methylated rNpm (Rme rNpm) in egg extract and oocyte extract supplemented with γ-32P-ATP; shown are autoradiogram (top) and Coomassie stained gel (bottom). The weak band appearing in the Npm negative lane in the egg extract is the endogenous Npm from the extract. F. Npm domains with identified sites of modification (“P”, phosphorylation; “EE”, glutamylation; “Rme”, arginine methylation) shown above. G. Our inferred biological order of events during oogenesis. H. A model summarizing our results. Shown are the N-terminal tail (blue box with white outline), A2 (red box), and C-terminal tail (thick blue line) with representative PTMs for each stage. Also See Figure S7.