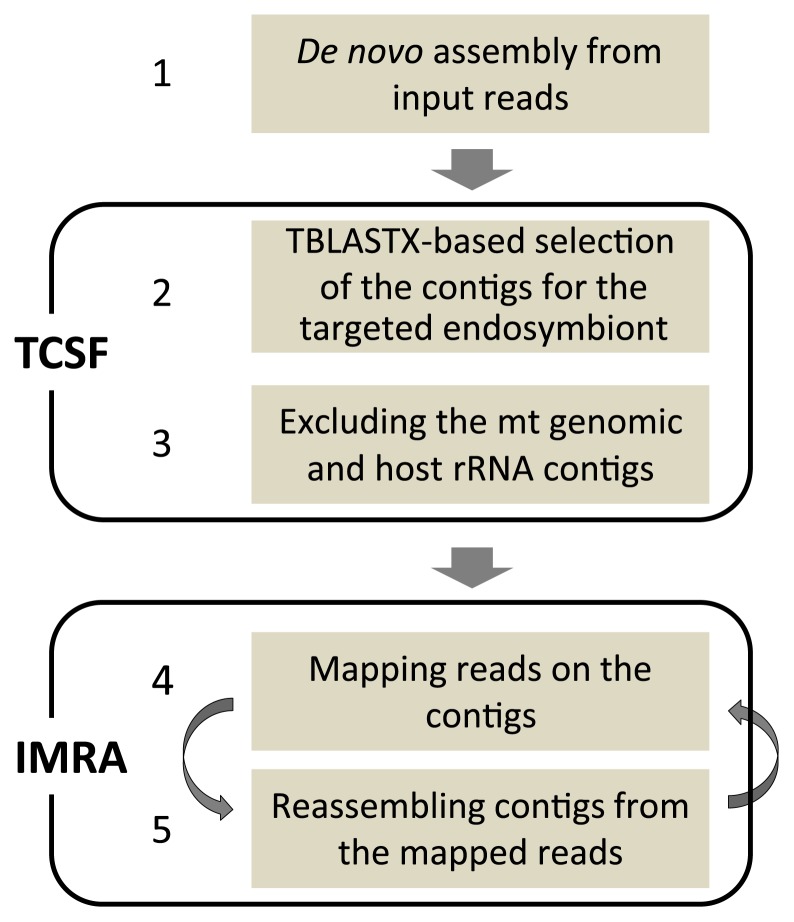
Fig. 1.
Flowchart of the sequence-assembly procedure developed in this study. (1) De novo sequence assembling from input reads, performed in the paired-end mode by using Roche Newbler software. (2) Selecting contigs for endosymbionts by performing TBLASTX searches (E<10−12) against the genomic sequence of a related genome. (3) Filtering the selected contigs by using BLAST to exclude contigs obtained from mitochondrial genomes (i.e. contigs with higher scores to mitochondrial genomes than to the related genome by using TBLASTX, E<10−12) and rRNA genes derived from the host genome (by using BLASTN, E<10−12). (4) Mapping input reads onto the filtered contigs by using the Roche Reference Mapper software in the single-end mode. (5) Reassembling contigs from the mapped reads by using the paired-end mode (minimal length of contigs: 500 nt). Iterations of (4) and (5) are continued until the increase in the total length of contigs and the reduction in the number of contigs reach saturation.