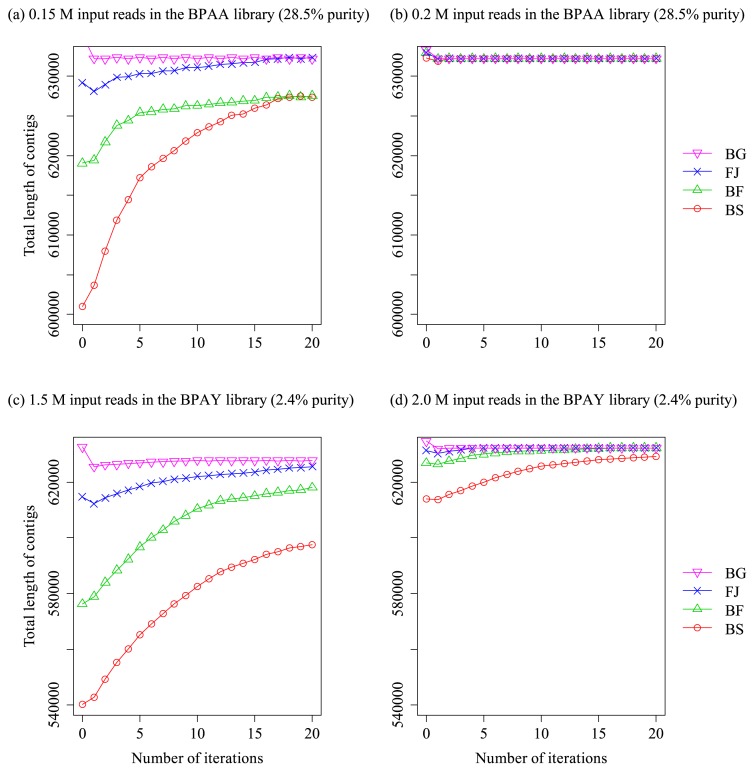
Fig. 4.
Total length of contigs with increasing iterations in the IMRA procedure used with distinct combinations of target and reference genomes. The IMRA procedure was started with filtered contigs obtained following the use of the TCSF procedure with the reference genome. Target endosymbiont genomes and the number of input reads are as follows: (a) B. cuenoti strain BPAA (28.5% purity in the DNA library) with 0.15 M input reads; (b) B. cuenoti strain BPAA (28.5% purity) with 0.2 M input reads; (c) B. cuenoti strain BPAY (2.4% purity) with 1.5 M input reads; and (d) B. cuenoti strain BPAY (2.4% purity) with 2 M input reads. Abbreviations for reference genomes used in the preceding TCSF procedure: BG, B. cuenoti strain BGIGA; FJ, Flavobacterium johnsoniae; BF, Bacteroides fragilis; BS, Bacillus subtilis.