Summary
The mammalian gastrointestinal (GI) microbiota is highly adapted to thrive in the GI environment and performs key functions related to host nutrition, physiology, development, immunity, and behavior. Successful host-bacterial associations require chemical signaling and optimal nutrient utilization and exchange. However, this important balance can be severely disrupted by environmental stimuli, with one of most common insults upon the microbiota being infectious diseases. Although the microbiota acts as a barrier towards enteric pathogens, many enteric pathogens exploit signals and nutrients derived from both the microbiota and host to regulate their virulence programs. Here we review several signaling and nutrient recognition systems employed by GI pathogens to regulate growth and virulence. We discuss how shifts in the microbiota composition change host susceptibility to infection, and how dietary changes or manipulation of the microbiota could potentially prevent and/or ameliorate GI infections.
Introduction
The mammalian GI tract is a complex environment where bacterial-host associations are paramount. The gut is populated by a dense and diverse microbiota that is intrinsically connected to host health and disease states. It is now appreciated that in addition to providing nutrients and vitamins, the microbiota impacts the host’s metabolism, immune and digestive systems, behavior, and neurological diseases (Ferreyra et al., 2014a; Hooper et al., 2002; Sampson and Mazmanian, 2015; Sharon et al., 2014; Sommer and Backhed, 2013). Moreover, the microbiota has been historically regarded as a barrier to enteric pathogens, commonly referred to as colonization resistance (Bohnhoff et al., 1954; Mushin and Dubos, 1965). These diverse roles of the gut microbiota have to be coordinated amongst the hundreds of different bacterial species and the host itself. This coordination is achieved through an array of chemicals that range from signaling molecules to metabolites and several that moonlight in both roles. The chemistry within the intestine is diverse, but still poorly understood (Marcobal et al., 2013). Many enteric pathogens exploit this intestinal chemistry to recognize the environment and gauge host physiology. These pathogens are crafty in recognizing different microbiota as well as host derived signals and nutrients to coordinate expression of their virulence traits, and adjust their metabolism to ensure successful competition for nutrients and a colonization niche (Hughes and Sperandio, 2008; Pifer and Sperandio, 2014).
Bacterial pathogens sense these chemicals through diverse receptors that generally are themselves transcription factors, or relay this information to transcription factors. Many of these signals and/or nutrients are sensed by membrane-bound histidine sensor kinases (HKs) that increase their phosphorylation in response to these signals, and initiate an intracellular signaling cascade within the bacterial cell, where the kinase transfers the phosphate to a response regulator (RR) that is activated upon phosphorylation. The vast majority of the RRs are transcription factors that regulate expression of different sets of genes, coordinating the response of the bacterial cell to certain environmental cues. Together, the HK and the RR comprise a two component signaling (TCS) system. In addition to TCSs, bacterial cells also recognize signals/nutrients through intracellular receptors that are themselves transcription factors. Upon binding their specific chemical ligands, these intracellular receptors change their conformation either increasing or decreasing their affinity to DNA to modify gene expression (Sperandio and Freitag, 2012).
These signaling systems often regulate a vast array of virulence factors that determine the success of an infection. Enteric pathogens have different adhesins to promote adherence to epithelial cells, specialized secretion systems such as the syringe-like type three secretion systems (T3SSs) to translocate bacterial effectors to host cells to highjack their function, as well as toxins that can either change signal transduction or kill eukaryotic cells among many other virulence genes. The genes encoding these virulence factors have usually been horizontally acquired through transposition, conjugation or phage transduction, and are spatially clustered in pathogenicity islands (PAI) (Kaper and Hacker, 1999). Virulence genes are generally employed as competition tools by enteric pathogens to gain access to unique niches inaccessible to the microbiota (Kamada et al., 2012a), such as the interface with the intestinal epithelium, and to invade and survive within epithelial cells and macrophages. However, expression of this virulence repertoire can be energetically expensive, and dysregulated expression is onerous to the pathogen. To ensure the correct timing and niche for expression of these traits, pathogens survey the chemistry landscape within the gut.
In this review we examine how several enteric pathogens actually thrive with the “naïve” aid of certain members of the microbiota. They interpret signal and nutritional cues from both the microbiota and the host to assess competition for nutrients and colonization sites, coordinating virulence and metabolism to ensure optimal colonization of the host. We discuss the various signaling systems employed by these pathogens to recognize these cues, and entertain how differences in microbiota composition due to dietary changes, genetic conditions or antibiotic treatment may impact the course of enteric infections.
The microbiota and nutrient utilization in the gut
The GI tract contains a wide variety of nutrients from several different sources, both endogenous and exogenous (Koropatkin et al., 2012; Sonnenburg et al., 2005). The host produces a mucosal layer to protect the epithelia, consisting of heavily glycosylated mucin proteins. Mucosal glycans contain multiple different sugar residues (Larsson et al., 2009) that are important nutrient sources for the microbiota as well as intestinal pathogens. Carbohydrates, amino acids and other nutrients consumed in the host diet feed luminal bacterial populations, and are constantly changing based on host diet. Finally, different members of the microbiota produce a variety of metabolic byproducts that can be utilized by other species or influence their physiology. The nutrients present in the gut have profound effects on the composition of the bacterial community, but also act as signals to influence the physiology of both commensal and invading pathogenic microbes.
Microbiota generation of nutrients
The GI tract is home to hundreds of bacterial species, all surviving on a finite number of resources. This gives rise to an incredibly complex food web where competition, cooperation and synergism all occur. In order to expand in this saturated environment, incoming pathogens must exploit portions of this nutritional web to proliferate and successfully deploy their virulence regime.
Certain members of the microbiota, most notably members of the phylum Bacteroidetes, have a greatly expanded ability to degrade complex carbohydrates (El Kaoutari et al., 2013). Other species that lack the requisite enzymes to degrade these large glycan structures can take advantage of sugars released during this breakdown process as a nutrient source. Several recent studies have demonstrated that microbiota-liberated sugars are important nutrients for enteric pathogens when establishing an infection. The ability to catabolize sialic acid, a sugar found in the mucus layer, increases colonization levels of both Clostridium difficile and Salmonella typhimurium in an antibiotic treated mouse model, despite both of these organisms lacking a sialidase enzyme to liberate this sugar. Instead, these two pathogens depend on the sialidase activity of the microbiota and an abundant commensal species, Bacteroides thetaiotaomicron [B. theta], could restore the advantage of sialic acid catabolism in germ free mice (Ng et al., 2013). Interestingly, B. theta does not have a catabolic pathway for sialic acid (Xu et al., 2003) and presumably encodes this enzymatic activity solely to access the underlying sugars in mucosal glycans, which are a significant nutrient source for this organism (Martens et al., 2008). Similarly, vancomycin-resistant Enterococcus, which cannot grow on purified mucin, can grow on mucin pre-digested with extracts from human stools, suggesting that it can take advantage of microbiota-liberated mucosal sugars in the gut (Pultz et al., 2006). These data demonstrate how diverse pathogens with different lifestyles have evolved a common strategy for co-opting the glycosidic abilities of commensal microbes to expand in the gut.
In addition to affecting the availability of sugars through enzymatic activity, the microbiota can also directly influence the production of mucosal glycans by the host. Probiotic Lactobacillus species can increase expression of mucins by intestinal epithelial cells (Mack et al., 1999), altering the carbohydrate landscape of the mucosal layer. The microbiota is also required for fucosylation of mucosal glycans, as fucose residues are not found in the mucus layer of germ-free mice. Colonization with B. theta can rescue this phenotype, but is dependent on B. theta’s ability to catabolize fucose (Bry et al., 1996; Hooper et al., 1999).
In addition to affecting the availability of sugars, the microbiota produces a wide variety of metabolic by-products such as gases, short chain fatty acids and organic acids that can be utilized by enteric pathogens as nutrients. C. difficile relies on succinate, an organic acid made by many members of the gut microbiota as a by-product of anaerobic fermentation (Reichardt et al., 2014). C. difficile utilizes succinate as an electron sink, converting it to butyrate to regenerate NAD+ (Ferreyra et al., 2014b). This allows fermentation of dietary sugars like sorbitol, a sugar alcohol whose levels increase following antibiotic treatment [a major risk factor for C. difficile infection] (Theriot et al., 2014). The succinate to butyrate pathway confers a growth advantage to C. difficile in vivo (Ferreyra et al., 2014b), suggesting that C. difficile has evolved to take advantage of the nutrients in the post-antibiotic gut. Molecular hydrogen is another abundant by-product of anaerobic fermentation by the microbiota. The hyb hydrogenase allows S. typhimurium to utilize microbiota-produced hydrogen as an energy source and enhances its growth during the initial invasion stage of infection. This is dependent on the microbiota as there is no advantage associated with hyb in germ-free mice (Maier et al., 2013). For pathogens entering the densely populated gut, co-opting molecules produced by resident microbes is an important strategy for initial expansion.
Pathogens create a distinct nutrient niche
Although the gut environment contains a wide variety of nutrients, the immense number of microbes creates intense competition for resources. One mechanism to compete with the microbiota is for pathogens to evolve a distinct metabolic repertoire. In a streptomycin-treated mouse model Enterohemorrhagic E. coli (EHEC) EDL933 and commensal E. coli strains MG1655, Nissle 1917 and HS differ in which sugars they use to establish and maintain colonization. Mutants defective in catabolic pathways for each of the 12 sugars found in the mucus layer revealed that each E. coli strain required a unique set of sugars for full colonization (Fabich et al., 2008b; Maltby et al., 2013). Although EDL933 could colonize the intestines of mice that were pre-colonized with any of the commensal strains alone, pre-colonization with a combination of the three strains prevented EDL933 colonization (Leatham et al., 2009). However, the sugar utilization profile of E. coli Nissle 1917 and HS together covers all of the sugars important for MG1655 colonization and accordingly, these two stains protect equally well as the three strain cocktail against EDL933 colonization (Maltby et al., 2013). This provides evidence for a nutrient competition mechanism of colonization resistance against pathogenic E. coli by closely related strains. Notably, catabolic pathways for two substrates, hexuronates [glucuronate and galacturonate] and sucrose, are important for EDL933 colonization but are either not present or not required for colonization by E. coli MG1655, Nissle 1917 or HS (Fabich et al., 2008b; Maltby et al., 2013), suggesting that pathogenic E. coli has evolved partially distinct nutrient requirements from its commensal relatives.
Interestingly, the colonization defects observed for pathogenic EDL933 upon loss of multiple catabolic pathways are additive, suggesting this strain metabolizes multiple sugars simultaneously. This phenomenon was not observed for commensal MG1655, suggesting it utilizes available sugars in a stepwise hierarchy (Fabich et al., 2008b). This supports the idea that not only has pathogenic E. coli evolved to utilize distinct nutrients from commensal strains, but also employs a different metabolic strategy. This hypothesis is further supported by the fact that EDL933 switches from using glycolytic to gluconeogenic substrates in a mouse gut pre-colonized with either MG1655 or Nissle 1917, and an EDL933 mutant unable to utilize gluconeogenic substrates cannot expand to or sustain wild-type colonization levels under these conditions (Miranda et al., 2004a; Schinner et al., 2015). This demonstrates that in order to compete with closely related commensal species that are highly adapted for the gut, enteric pathogens have evolved unique metabolic profiles and strategies.
Competition for nutrients occurs between more distantly related species as well. Citrobacter rodentium, a mouse pathogen used as a model for EHEC disease, downregulates its virulence genes by 21 days post-infection and is then outcompeted by the microbiota and cleared from the mouse gut. However, this phenomenon was shown to be dependent on nutrient availability in a B. theta colonized gnotobiotic mouse model. When mice are fed a diet containing both monosaccharides, which can be utilized by Enterobacteriacae like C. rodentium, and polysaccharides, which can be utilized by Bacteroides species, C. rodentium is able to colonize and cause disease. However, when the mice are switched to a diet containing only monosaccharides and the two species are forced to compete for sugars, B. theta out-competes C. rodentium and the pathogen is cleared (Kamada et al., 2012b). Establishing a unique metabolic niche is crucial for invading pathogens to be able to compete with commensal species and expand (Fig.2).
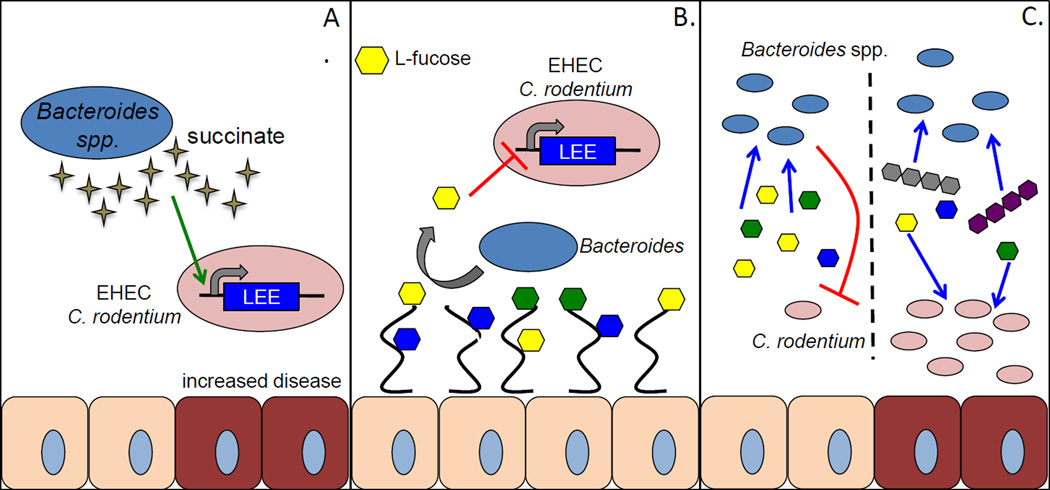
Fig. 2.
Modulation of virulence by commensal Bacteroides. Commensal Bacteroides affect virulence and progression of disease by AE pathogens in several ways. A. Bacteroides produce a significant amount of succinate as a by-product of carbohydrate fermentation. Succinate is sensed by EHEC and C. rodentium and upregulates expression of the LEE PAI. B. L-fucose is liberated from host mucin glycoproteins by fucosidases expressed by members of the microbiota such as Bacteroides thetaiotaomicron. Free fucose is sensed by EHEC and represses the LEE PAI to prevent early activation of this virulence factor before reaching the epithelium. C. Nutrient competition between Bacteroides spp. and pathogenic enterobacteriaceae significantly affects the progression of disease. When only simple sugars (mono and di-saccharides) are present commensal Bacteroides and pathogens are forced to compete for nutrients, which limits growth and eventually leads to clearance of the pathogen. When both simple sugars and complex carbohydrates are present Bacteroides will preferentially utilize polysaccharides, and pathogenic enterobacteriacae are able to utilize simple sugars to proliferate and persist in the intestine.
Usage of ethanolamine also allows pathogens a nutrient source distinct from competing commensals. Ethanolamine is a component of an abundant phospholipid in mammalian and bacterial membranes and is found in the intestinal tract due to epithelial cell turnover (Bertin et al., 2011). Ethanolamine can be used as a carbon and/or nitrogen source by several intestinal pathogens (Garsin, 2010), and in fact ethanolamine utilization [eut] genes are preferentially found in the genomes of bacteria that cause food borne illness (Korbel et al., 2005). In the intestine this metabolite can serve as a selective nutrient source for pathogens as the majority of commensal species do not encode eut genes. Ethanolamine utilization by S. typhimurium and EHEC provides a competitive growth advantage in the intestinal tract (Bertin et al., 2011; Thiennimitr et al., 2011) and in Listeria monocytogenes it enhances intracellular replication (Joseph et al., 2006). Expression of the eut operon is modulated by global regulators of virulence in S. typhimurium, Enterococcus faecalis and L. monocytogenes, suggesting that ethanolamine utilization is tied to virulence (Bourgogne et al., 2006; Kelly et al., 2004; Lawhon et al., 2003; Toledo-Arana et al., 2009). Indeed, ethanolamine activates virulence gene expression in EHEC by binding to the EutR transcription factor, demonstrating that it can be both a nutrient source and a signaling molecule (Gonyar and Kendall, 2014; Kendall et al., 2012; Luzader et al., 2013).
This scavenging of nutrients is critical for establishing colonization in the gut and requires the utilization of unique nutrient sources in order to effectively compete with the hundreds of commensal bacteria that occupy and proliferate within the GI tract. Beyond providing these basic growth requirements, metabolism and virulence are intimately intertwined and many pathogens have evolved to recognize metabolite cues as signals to regulate virulence genes (Fig. 1).
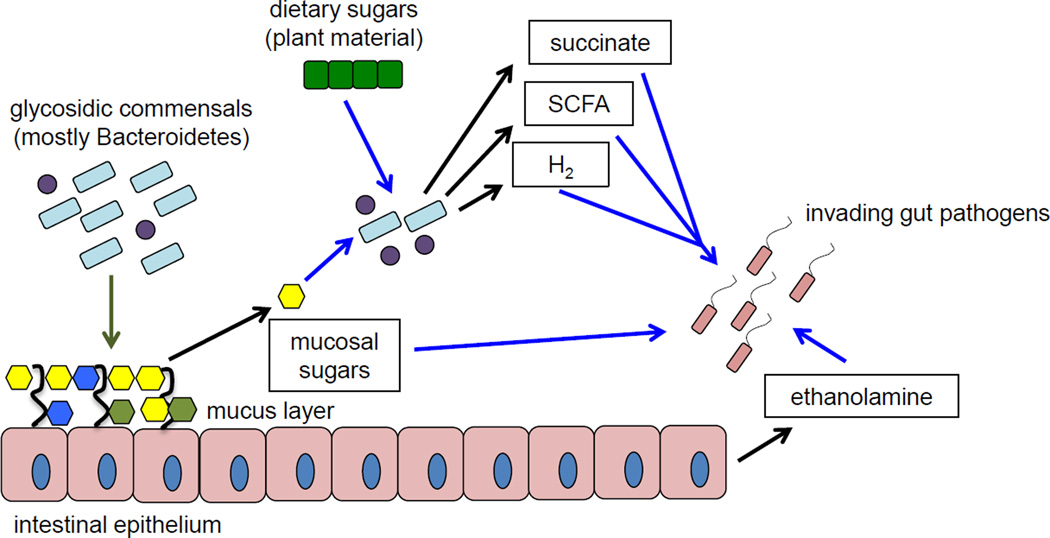
Fig 1.
Microbiota-derived nutrients feed pathogenic bacteria. Black arrows indicate production of a particular nutrient, blue arrows indicate consumption of the indicated nutrient. Members of the intestinal microbiota stimulate (green arrow) mucosal sugar production by the host as well as produce glycosidic enzymes that liberate mucosal sugars (galactose, fucose, sialic acid etc) from host mucin glycoproteins. Liberated mucosal sugars can directly feed invading pathogen populations. Fermentation of dietary and host-derived sugars by the microbiota leads to production of SCFA, hydrogen and organic acids like succinate, which can also serve as nutrient sources for pathogens during infection. Epithelial cell turnover releases ethanolamine into the lumen of the gut, where it can serve as a selective nutrient for pathogen proliferation during inflammation.
Microbiota derived metabolites as virulence signals
Obtaining nutrients and proliferating is the first crucial step for pathogens to establish a productive infection. However, after proliferating to sufficient levels pathogens must precisely regulate when and where they will deploy their virulence program. Microbiota-generated metabolites can act as signals that regulate the production of virulence factors and ultimately affect the progression of disease (Fig. 1 and 2).
The nutritional state of the gut [nutrient rich vs nutrient poor], which is profoundly affected by the microbiota, can be a signal for inducing or repressing virulence genes. EHEC preferentially activates expression of its locus of enterocyte effacement (LEE) PAI in gluconeogenic vs. glycolytic conditions. The LEE encodes for a T3SS, an adhesin and effectors that are essential for EHEC to form attaching and effacing (AE) lesions on enterocytes (Kaper et al., 2004). In the presence of low glucose levels or the gluconeogenic substrate succinate, LEE transcription levels are higher than when glucose is abundant. This regulation occurs through the concerted effort of two transcriptional regulators Cra and KdpE (Njoroge et al., 2012). Cra is an intracellular transcription factor, which is a master regulator of carbon metabolism that represses transcription of glycolytic enzymes and activates gluconeogenic enzymes (Saier and Ramseier, 1996b). Cra senses fluctuations in sugar concentrations to modulate its function (Ramseier et al., 1993). When E. coli is growing in the presence of glycolytic substrates, there is accumulation of fructose-1-phosphate (F1P) and fructose-1,6-bisphosphate (FBP), which bind to Cra decreasing its binding affinity to DNA, consequently decreasing its regulatory function, including LEE transcription in EHEC (Njoroge et al., 2012; Ramseier et al., 1995; Saier and Ramseier, 1996a). KdpE is the RR of the KdpDE two-component system that regulates various genes in response to osmotic stress (Jung et al., 1997). However, both of these regulators have been co-opted in pathogenic E. coli to regulate virulence. Under gluconeogenic conditions Cra and KdpE interact while binding to different regions of the ler promoter (that encodes the Ler transcription factor that activates expression of all of the LEE genes (Mellies et al., 1999)), causing increased expression of the LEE genes involved in T3SS and increasing AE lesion formation (Njoroge et al., 2012). Cra and KdpE also regulate several other non-LEE encoded virulence factors, either together or individually, in response to a low glucose conditions (Njoroge et al., 2013). Shigella flexneri also regulates virulence genes in response to nutrient conditions, where glycolysis is tied to virulence gene expression. S. flexneri mutants inhibited for glycolysis at various stages display decreased expression of the virF and virB virulence regulators causing a loss of invasion plasmid antigen expression and decreased cellular attachment and invasion (Gore and Payne, 2010) (Figs. 2 and 3).
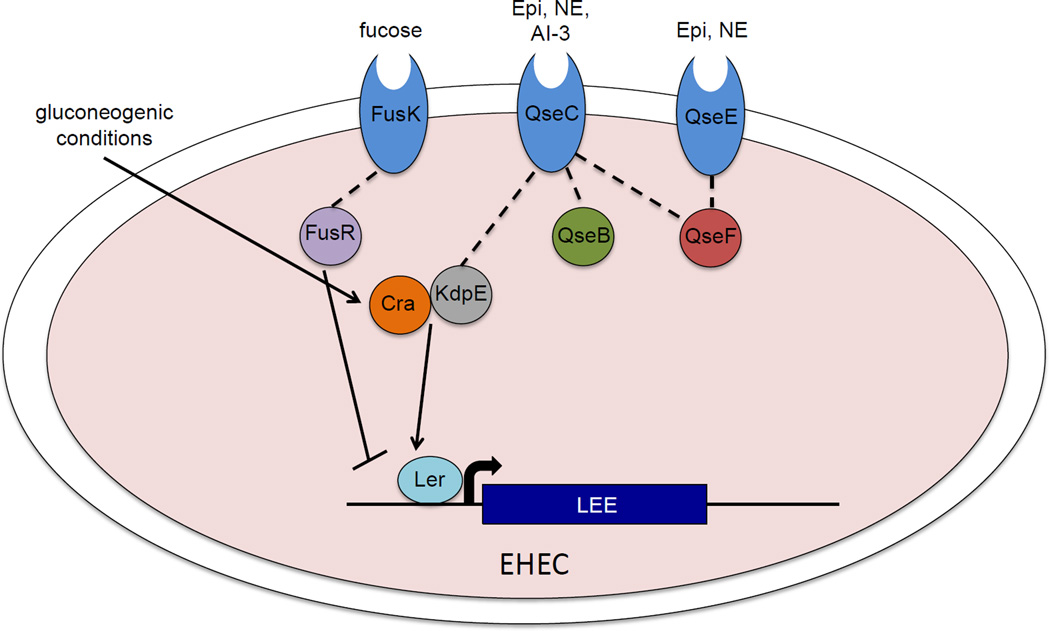
Fig. 3.
Adrenergic and nutrient signals intersect to control expression of the EHEC LEE. Host hormones Epinephrine (Epi) and Norepinephrine (NE), whose intestinal availability is modulated by the microbiota, are recognized by two sensor histidine kinases, QseC and QseE. QseC also recognizes the microbiota-derived quorum sensing molecule AI-3. QseC and QseE phosphorylate their cognate response regulators (RR) QseB and QseE respectively and QseC also phosphorylates RRs KdpE and QseF. KdpE interacts with the master regulator of carbon metabolism Cra to activate Ler, which Cra also activates under gluconeogenic conditions (ex: low glucose levels, high levels of succinate). The mucosal sugar fucose, whose liberation is microbiota dependent, is recognized by another HK FusK. FusR, the cognate RR of FusK, represses activation of the LEE. Expression of FusKR is repressed by the QseC/QseE signalling cascade.
Microbiota-produced metabolites can also be specifically sensed by pathogens as signals to induce or repress virulence genes. In a C. rodentium infection model, antibiotic-treated mice that are reconstituted with high levels of B. theta before being infected display more severe pathology and succumbed to disease more quickly than mice that were left depleted of their microbiota. Metabolomics revealed that succinate levels are significantly higher in the B. theta reconstituted mice compared to non-reconstituted mice (Curtis et al., 2014b). Additionally, succinate stimulates EHEC production of the T3SS component EspA in vitro. This succinate response is mediated through the carbon metabolism master regulator Cra (Curtis et al., 2014b), suggesting that EHEC and C. rodentium sense the metabolic environment of the gut through microbiota-produced metabolites and regulate their virulence genes accordingly.
Aside from being indicators of the energy state of a particular environment, metabolites can also be used as cues to determine precise location within the gut, which is a critical factor for determining whether to express virulence genes. EHEC encodes the FusKR TCS that senses the mucosal sugar fucose and represses expression of the LEE PAI. The genes encoding fusKR have been recently horizontally acquired and are within the PAI OI-20 (Pacheco et al., 2012). The fusKR genes were acquired by EPEC O55:H7 that gave rise to EHEC O157:H7 (Reid et al., 2000; Wick et al., 2005). This TCS is also present in C. rodentium, suggesting that it is exclusively found in AE GI pathogens that colonize the colon. It is noteworthy that FusKR is not present in other commensal or pathogenic E. coli strains, or enteric bacteria at large, and homologs are only found in Enterococcus fecaelis, suggesting that EPEC O55:H7 acquired fusKR from E. fecaelis, and later evolved into EHEC O157:H7. OI-20 genes are up-regulated when EHEC is grown in the presence of mucus (Bai et al., 2011), and during infection of the colonic mucus-producing cell line HT29, suggesting that expression of this TCS in mucus facilitates EHEC adaptation to the mammalian intestine (Pacheco et al., 2012). Thus, it is tempting to speculate that acquisition of OI-20 enhances EHEC’s capability to successfully compete for a niche in the colon.
Generation of the fucose signal is dependent on the microbiota as several members of the microbiota, but not EHEC, have the enzymatic capacity to cleave the sugar residues from mucin glycoproteins. It is hypothesized that LEE repression by fucose is to prevent early expression of the energetically expensive T3SS, and allow expression only once the pathogen has passed through the mucosal layer to the host epithelium (Pacheco et al., 2012). FusR encodes a RR that directly represses expression of the LEE genes, by repressing transcription of ler. FusK, the HK of this TCS, autophosphorylates in response to fucose, thus revealing a signal transduction mechanism that senses fucose to regulate expression of the LEE as well as EHEC intestinal colonization in the infant rabbit model of infection. In addition to LEE regulation, FusKR also indirectly represses expression of the fuc genes involved in fucose utilization through regulation of the Z0461 hexose-phosphate-major facilitator-superfamily (MFS) transporter, also encoded within the OI-20 (Pacheco et al., 2012). EHEC competes with commensal E.coli, but not B.theta, for the same carbon sources (e.g. fucose) within the mammalian intestine (Autieri et al., 2007; Chang et al., 2004; Fabich et al., 2008a; Fox et al., 2009; Kamada et al., 2012a; Miranda et al., 2004b). Commensal E.coli, however, are not found in close contact with the epithelia, being in the mucus-layer, where it is counter-productive for EHEC to invest resources to utilize fucose, when EHEC can efficiently use other carbon sources such as galactose, hexorunates, and mannose, which are not used by commensal E.coli within the intestine (Fabich et al., 2008a). Additionally, in contrast to commensal E.coli, EHEC is found closely associated with the intestinal epithelium (Miranda et al., 2004b). Therefore, EHEC can utilize nutrients exclusively available at the surface of the epithelial cells. Consequently, the decreased expression of the fuc operon through fucose-sensing by FusKR, may prevent EHEC from expending energy in fucose utilization in the mucus-layer, where it competes with commensal E.coli for this resource, and focus on utilizing other carbon sources (e.g. galactose, whose utilization is not affected by FusKR, not used by this competitor. Thus, the colonization defect of ΔfusK of the mammalian GI tract results from its inability to correctly time virulence and metabolic gene expression (Pacheco et al., 2012) (Figs 2 and 3).
Short chain fatty acids [SCFA] are some of the most abundant fermentation by-products produced by the microbiota in the intestinal tract. The most abundant three are acetate, propionate and butyrate but the concentrations and proportions of these molecules differ along the length and width of the gut. Therefore, some enteric pathogens have evolved to sense SCFAs as biomarkers of their location in the gut. Mixtures of acetate, propionate and butyrate in concentrations and proportions that mimic the ileum enhance expression of the Salmonella pathogenicity island 1 (SPI-1) by S. typhimurium where colonic-like SCFA mixtures suppress SPI-1 expression and inhibit cell invasion (Lawhon et al., 2002). This reflects S. typhimiurium’s preference for the ileum as the primary site for cell invasion (Carter and Collins, 1974). More specifically, exposure to acetate alone enhances SPI-1 expression (Lawhon et al., 2002) while exposure to butyrate and propionate decreases SPI-1 expression levels (Gantois et al., 2006; Hung et al., 2013). The mechanism of SPI-1 regulation by SCFA is not completely understood but it appears that the three SCFAs affect different parts of the SPI-1 regulatory cascade upstream of the HilA master regulator. Additionally, propionate and acetate must be internalized and converted to propionyl-CoA and acetyl-CoA respectively to exert their effects (Hung et al., 2013; Lawhon et al., 2002). In contrast to Salmonella, EHEC deploys its virulence genes primarily in the colon, and accordingly the effects of the different SCFAs on virulence are reversed. In EHEC exposure to butyrate increases expression of the LEE PAI and enhances cell adherence, where exposure to acetate and propionate had little to no effect on virulence gene expression. This regulation is achieved through the Lrp transcriptional regulator, which is post-transcriptionally activated in the presence of butyrate and initiates a regulatory network involving several proteins that act cooperatively to enhance and prolong LEE expression (Nakanishi et al., 2009; Takao et al., 2014; Tobe et al., 2011). Exposure to SCFAs, especially butyrate, also increases expression of EHEC flagella through the action of Lrp as well as a regulatory cascade independent of that used to activate the LEE (Tobe et al., 2011) (Fig. 2).
Shifts in the microbiota affect intestinal pathogens
The microbiota is a dynamic community and its composition is influenced by several factors including diet, age, antibiotic use, disease state and others. These factors can influence not only the membership and relative abundance of the species composing the microbiota, but also their physiology, leading to different metabolite profiles. These microbiota metabolite shifts can have significant effects on host physiology as well as the physiology of invading microbes, ultimately affecting disease progression.
The composition of the microbiota has recently been recognized as a central factor in determining susceptibility to enteric infection and recent studies have demonstrated that this community can be manipulated to affect disease outcome. Distinct phylogenetic microbiota compositions are observed between different strains of mice and correspondingly different mouse strains naturally vary in their susceptibly to enteric infection. While some of this can be attributed to host genetics (Marquis and Gros, 2008), recent studies demonstrate that differences in the microbiota contribute significantly to the discrepancy between strains. Transferring the microbiota of a susceptible mouse to a resistant mouse, via antibiotic treatment and reconstitution, increases pathogen loads and pathology associated with C. rodentium infection. The reciprocal is also true where transplantation of a resistant microbiota will confer some protection to a genotypically susceptible mouse. Mice with a resistant microbiota [naturally or via transplant] display higher levels of pro-inflammatory cytokines, particularly IL-22, which aid in clearance of C. rodentium and enhance survival (Ghosh et al., 2011; Willing et al., 2011). Currently, a similar microbiota transplantation technique is being used clinically to treat human C. difficile infection [CDI]. CDI is often preceded by antibiotic use or another disturbance in the normal composition of the patient’s microbiota that decreases the diversity of the population (Chang et al., 2008). Following transplantation from a healthy donor, diversity of the community begins to rebound and the community structure resembles that of the donor (Hamilton et al., 2013; Song et al., 2013). This shift in microbiota composition corresponds with a successful recovery in CDI patients receiving microbiota transplant (Khoruts et al., 2010), demonstrating how the community can be manipulated to successfully treat disease.
Antibiotic use has long been known to be a risk factor for infection by enteric pathogens (Pavia et al., 1990; Pepin et al., 2005). Research over the past several years has revealed that one of the mechanisms by which antibiotics decrease colonization resistance by the microbiota is by shifting the metabolic environment in a way that pathogens can exploit. Metabolomics revealed numerous differences in the mouse intestine before and after antibiotic treatment that correlated with susceptibility to C. dificile infection. The metabolite landscape before antibiotic treatment was more similar to mice allowed to recover from treatment for 6 weeks [both resistant states] than to mice directly following antibiotic treatment [susceptible state] (Theriot et al., 2014). Following antibiotic treatment there is an increase in molecules that support C. difficile germination and growth. Metabolites like sialic acid, sorbitol and succinate that provide C. difficile with a growth advantage in the gut, all transiently increase (Ferreyra et al., 2014b; Ng et al., 2013; Theriot et al., 2014). Increased levels of taurocholate, a bile acid that enhances C. difficile spore germination, are also observed after antibiotic treatment (Theriot et al., 2014).
Metabolic shifts associated with changes in the microbiota can also alter how the host is affected by certain virulence factors. Exposure to butyrate enhances host cell expression of globotriaosylceramide [Gb3], the receptor for Shiga toxin, a cytotoxin common to E. coli and Shigella. When mice are fed a high fiber diet the resulting increase in butyrate output by the microbiota correlates with more severe pathology and faster death during EHEC infection than mice on a low fiber diet that have lower intestinal butyrate levels (Zumbrun et al., 2013). It is hypothesized that the shift in diet alters the microbiota such that more butyrate is being produced, which increases Gb3 expression on host cells and increases their susceptibility to Shiga toxin. Conversely, a microbiota composition that produces increased acetate levels can protect against Shiga toxin mediated disease. In a lethal model of EHEC infection, mice colonized with certain Bifidobacteria that lead to higher acetate levels in the gut get less severe disease compared to mice associated with Bifidobacteria strains that lead to lower acetate levels. Microbiota-produced acetate increases barrier function of the intestinal epithelium and prevents Shiga toxin from passing into the blood stream (Fukuda et al., 2011). The composition and metabolism of the microbiota is a critical factor in determining susceptibility to and progression of disease, and represents an exciting new target for preventing and treating enteric infection.
Integration of nutritional and adrenergic signaling in the gut
The GI tract is highly innervated, and neurotransmitters are prominent in the GI environment. These neurotransmitters have important physiological functions in the gut where they modulate intestinal smooth muscle contraction, submucosal blood flow, and chloride and potassium secretion (Horger et al., 1998). There is also an important relationship between neurotransmitters and the microbiota. The microbiota induces biosynthesis of serotonin (Yano et al., 2015), and modulates the levels of the stress hormones epinephrine and norepinephrine (NE) in the gut lumen (Asano et al., 2012). Epinephrine and NE are at the core of stress responses (Molina, 2006), and an important chemical exchange within the gut involves these neurotransmitters. Both of them are present in the gut, with NE being synthesized by the adrenergic neurons within the enteric nervous system (ENS) (Furness, 2000). Epinephrine is synthesized in the central nervous system (CNS) and in the adrenal medulla; but it can reach the gut through the bloodstream (Purves et al., 2001). Stress has profound effects in GI function leading to increased gastric acid production and intestinal motility, and has also been shown to alter the composition of the gut microbiota in animals subjected to premature separation from their mothers (Grenham et al., 2011). Importantly epinephrine and NE can be detected in the lumen of the gut in their free active form. The luminal levels of NE increase from the ileum to the colon, with the higher concentrations being in the colon. The microbiota plays a critical role in the availability of active NE in the lumen. The host conjugates NE to glucuronic acid (glucuronide) to inactivate it. The microbiota encodes glucuronidases that deconjugate glucuronic acid from NE, increasing the levels of free biologically active NE in the lumen. In the lumen of germ free mice there is decreased free NE, with the majority of it being in the glucuronide inactive form (Asano et al., 2012).
There is an extensive body or work showing that epinephrine and NE increase virulence of several GI pathogens such as EHEC, S. typhimurium, and Vibrio parahaemolyticus (Curtis and Sperandio, 2011; Moreira et al., 2010; Nakano et al., 2007; Sperandio et al., 2003a). The epinephrine/NE signaling cascade has been elucidated in more detail in EHEC, where it increases expression of the LEE, Shiga toxin, flagella and motility (Hughes et al., 2009; Sperandio et al., 2003b). There are two bacterial adrenergic receptors, QseC and QseE, which are HKs that increase their autophosphorylation upon binding to epinephrine or NE (Clarke et al., 2006; Reading et al., 2009). QseC phosphorylates its cognate RR QseB, and the non-cognate RRs KdpE and QseF, while QseE exclusively phosphorylate its cognate RR QseF (Hughes et al., 2009; Reading et al., 2009; Yamamoto et al., 2005). The concerted action of this signaling cascade increases virulence gene expression in EHEC, but also leads to profound metabolic changes that allow EHEC to successfully colonize the colon (Curtis et al., 2014c; Hughes et al., 2009; Njoroge and Sperandio, 2012; Pacheco et al., 2012; Rasko et al., 2008). Importantly this bacterial adrenergic signaling cascade intertwines with sensing and integrating signals and nutrient cues provided by the gut microbiota. QseC, in addition to sensing epinephrine and NE, also senses autoinducer 3 (AI-3) that is a signal made by various members of the human microbiota, and is found in human stools (Sperandio et al., 2003b; Walters et al., 2006). Moreover, QseC phosphorylates KdpE that interacts with Cra, which senses succinate produced by B. theta, to activate LEE gene expression under gluconeogenic conditions that mirror the environment at the interface with the epithelium (Curtis et al., 2014a; Hughes et al., 2009; Njoroge et al., 2012). The QseBC and QseEF TCSs also repress expression of the FusKR system that senses fucose released from the mucus by B. theta to repress the LEE and adjust EHEC’s metabolism when it is in the outer mucus layer environment (Pacheco et al., 2012). This intricate relationship between adrenergic and nutrient signaling equips EHEC with a fine tuned signaling cascade to sense the colonic environment of the host, in addition to gaging whether it is in the lumen/outer mucus layer versus the interface of the epithelium (Fig. 3).
The survival of an organism lies within its intrinsic ability to detect and efficiently respond to stress cues. Stress responses play a key role in adaptation to environmental, psychosocial, and physical insults. Hence it comes as no surprise that stress responses require synchronization and coordination of an organism’s resources to ensure that metabolic substrates are available to meet the increasing energy demands of an effective stress response.
Trends for the future
As we are at the brink of appreciating the complex relationship between the host, gut microbiota and enteric pathogens, we are glancing at the exciting tip of an iceberg of knowledge. The rapid development of new technologies in genomics, metagenomics, metabolomics and transcriptomics are informing us about differential microbiota compositions due to different diets, host genetics and physiology. It is also clear that this relationship is a two way street, with microbiota metabolites greatly influencing host cell function. Moreover, the composition of the microbiota seems to determine host susceptibility to enteric pathogens. Some pathogens highjack microbiota and host derived signals and/or nutrients to promote disease and coordinate the expression of their virulence repertoire. Conversely, certain microbiota derived metabolites hamper virulence and infection. It is fascinating to ponder for example, why during an EHEC outbreak, in which the outbreak strain is the same, there is a whole range of differential symptoms and disease progression, with some having just watery diarrhea, others developing hemolytic colitis, and a few progressing to hemolytic uremic syndrome (HUS). There is also a distinct age susceptibility to HUS following an EHEC infection, with children generally under 5 years of age being at higher risk (Kaper et al., 2004). It is worthwhile considering that these differential disease progressions may have an important microbiota composition component to them. It has been well documented that one’s microbiota establishes itself around the age of 5 (Rodriguez et al., 2015), which coincides with the age range for enhanced susceptibility to EHEC-induced HUS.
Different microbiota derived metabolites have recently been shown to be sensed by EHEC to promote or decrease its virulence. Succinate, fucose, butyrate, and ethanolamine directly influence EHEC’s gene expression towards enhancement of virulence (Curtis et al., 2014a; Kendall et al., 2012; Nakanishi et al., 2009; Pacheco et al., 2012). High fiber diets increase the concentration of butyrate upregulating expression of the Shiga Toxin receptor on host cells enhancing susceptibility to HUS (Zumbrun et al., 2013). Meanwhile, acetate produced by Bifidobacteria decreases translocation of Shiga toxin through the intestine, protecting from HUS (Fukuda et al., 2011).
The metabolite exchange among host, microbiota and enteric pathogens, combined with the intrinsic relationship with adrenergic signaling opens a whole array of possibilities on prevention and/or amelioration/treatment of GI infections. It is tempting to speculate that several drugs commonly used to interfere with adrenergic signaling could be repurposed towards treatment of enteric infections. One could also foresee changes in diet or addition of supplements being explored as prebiotic therapy. Finally, there is the consideration of direct manipulation of the microbiota composition through pro-biotic approaches, or even stool transplants that have been so successful in treating C. difficile infections in humans.
Table 1.
Metabolites that contribute to pathogenesis
| A. Metabolites enhance pathogen expansion | |||
|---|---|---|---|
| Metabolite | Source | Pathogen | References |
| Sialic Acid | Released from mucosa by microbiota | S. typhimurium, C. difficile | Ng et al. 2013 |
| Succinate | By-product of microbiota fermentation | C. difficile | Ferreyra et al. 2014 |
| Hydrogen | By-product of microbiota fermentation | S. typhimurium | Maier et al. 2013 |
| Ethanolamine | Mammalian and bacterial membranes | EHEC, S. typhimurium, L. monocytogenes | Kendall et al. 2012; Bertin et al. 2011; Thiennimitr et al. 2011; Joseph et al. 2006 |
| Sorbitol | Host diet | C. difficile | Theriot et al. 2014 |
| B. Metabolites influence virulence gene expression | |||
|---|---|---|---|
| Metabolite | Virulence factor controlled |
Regulator(s) involved |
References |
| ethanolamine | Enhances LEE expression (EHEC) | EutR | Kendall et al. 2012 |
| Succinate | Enhances LEE expression (EHEC) | Cra | Curtis et al. 2014 |
| Fucose | Inhibits LEE expression (EHEC) | FusKR two component system | Pacheco et al. 2012 |
| Butyrate | Inhibits SPI-1 expression (S. typhimurium) | unknown | Lawhon et al. 2002; Gantois et al. 2006; Hung et al. 2013 |
| Enhances LEE expression (EHEC) | Lrp → PchA → LeuO → Ler | Nakanishi et al. 2009; Takao et al. 2014; Tobe et al. 2011 | |
| Enhances flagella expression (EHEC) | Lrp & unknown Lrp-independent pathway | Tobe et al. 2011 | |
| Propionate | Inhibits SPI-1 expression (S. typhimurium) | HilD → HilA | Lawhon et al. 2002; Gantois et al. 2006; Hung et al. 2013 |
| Acetate | Enhances SPI-1 expression (S. typhimurium) | BarA/SirA two component system → HilA | Lawhon et al. 2002 |
| Taurocholate | Enhances C. difficile germination | Theriot et al. 2014 | |
Acknowledgements
Work in the VS’s laboratory is supported by the National Institutes of Health grants AI053067, AI077613, AI101472, AI05135 and AI114511.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asano Y, Hiramoto T, Nishino R, Aiba Y, Kimura T, Yoshihara K, Koga Y, Sudo N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1288–G1295. doi: 10.1152/ajpgi.00341.2012. [DOI] [PubMed] [Google Scholar]
- Autieri SM, Lins JJ, Leatham MP, Laux DC, Conway T, Cohen PS. L-fucose stimulates utilization of D-ribose by Escherichia coli MG1655 DeltafucAO and E. coli Nissle 1917 DeltafucAO mutants in the mouse intestine and in M9 minimal medium. Infect Immun. 2007;75:5465–5475. doi: 10.1128/IAI.00822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, McAteer SP, Paxton E, Mahajan A, Gally DL, Tree JJ. Screening of an E. coli O157:H7 Bacterial Artificial Chromosome Library by Comparative Genomic Hybridization to Identify Genomic Regions Contributing to Growth in Bovine Gastrointestinal Mucus and Epithelial Cell Colonization. Front Microbiol. 2011;2:168. doi: 10.3389/fmicb.2011.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011;13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med. 1954;86:132–137. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]
- Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188:2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- Carter PB, Collins FM. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014a;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014b;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Russell R, Moreira CG, Adebesin AM, Wang C, Williams NS, Taussig R, Stewart D, Zimmern P, Lu B, et al. QseC inhibitors as an antivirulence approach for Gram-negative pathogens. MBio. 2014c;5:e02165. doi: 10.1128/mBio.02165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Sperandio V. A complex relationship: the interaction among symbiotic microbes, invading pathogens, and their mammalian host. Mucosal Immunol. 2011;4:133–138. doi: 10.1038/mi.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008a;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008b;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra JA, Ng KM, Sonnenburg JL. The Enteric Two-Step: nutritional strategies of bacterial pathogens within the gut. Cell Microbiol. 2014a;16:993–1003. doi: 10.1111/cmi.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014b;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JT, Drouillard JS, Shi X, Nagaraja TG. Effects of mucin and its carbohydrate constituents on Escherichia coli O157 growth in batch culture fermentations with ruminal or fecal microbial inoculum. J Anim Sci. 2009;87:1304–1313. doi: 10.2527/jas.2008-1166. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- Furness JB. Types of neurons in the enteric nervous system. Journal of the autonomic nervous system. 2000;81:87–96. doi: 10.1016/s0165-1838(00)00127-2. [DOI] [PubMed] [Google Scholar]
- Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, Hinton JC, Van Immerseel F. Butyrate specifically down-regulates salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol. 2006;72:946–949. doi: 10.1128/AEM.72.1.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Dai C, Brown K, Rajendiran E, Makarenko S, Baker J, Ma C, Halder S, Montero M, Ionescu VA, et al. Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am J Physiol Gastrointest Liver Physiol. 2011;301:G39–G49. doi: 10.1152/ajpgi.00509.2010. [DOI] [PubMed] [Google Scholar]
- Gonyar LA, Kendall MM. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2014;82:193–201. doi: 10.1128/IAI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore AL, Payne SM. CsrA and Cra influence Shigella flexneri pathogenesis. Infect Immun. 2010;78:4674–4682. doi: 10.1128/IAI.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Front Physiol. 2011;2:94. doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes. 2013;4:125–135. doi: 10.4161/gmic.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci U S A. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horger S, Schultheiss G, Diener M. Segment-specific effects of epinephrine on ion transport in the colon of the rat. Am J Physiol. 1998;275:G1367–G1376. doi: 10.1152/ajpgi.1998.275.6.G1367. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC) PLoS Pathog. 2009;5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DT, Sperandio V. Inter-kingdom signaling: communication between bacteria and host. Nature Reviews Microbiology. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, McClelland M, Ahmer BM, Altier C. The intestinal fatty acid propionate inhibits Salmonella invasion through the posttranslational control of HilD. Mol Microbiol. 2013;87:1045–1060. doi: 10.1111/mmi.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Tjaden B, Altendorf K. Purification, reconstitution, and characterization of KdpD, the turgor sensor of Escherichia coli. J Biol Chem. 1997;272:10847–10852. doi: 10.1074/jbc.272.16.10847. [DOI] [PubMed] [Google Scholar]
- Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science. 2012a;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012b;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Hacker J. Pathogenicity islands and other mobile virulence elements, first edn. Washington DC: ASM Press; 1999. [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kelly A, Goldberg MD, Carroll RK, Danino V, Hinton JC, Dorman CJ. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology. 2004;150:2037–2053. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- Kendall MM, Gruber CC, Parker CT, Sperandio V. Ethanolamine Controls Expression of Genes Encoding Components Involved in Interkingdom Signaling and Virulence in Enterohemorrhagic Escherichia coli O157:H7. MBio. 2012;3 doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Doerks T, Jensen LJ, Perez-Iratxeta C, Kaczanowski S, Hooper SD, Andrade MA, Bork P. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 2005;3:e134. doi: 10.1371/journal.pbio.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nature reviews Microbiology. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson JM, Karlsson H, Sjovall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19:756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol. 2003;48:1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x. [DOI] [PubMed] [Google Scholar]
- Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol. 2002;46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun. 2009;77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzader DH, Clark DE, Gonyar LA, Kendall MM. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2013;195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol. 1999;276:G941–G950. doi: 10.1152/ajpgi.1999.276.4.G941. [DOI] [PubMed] [Google Scholar]
- Maier L, Vyas R, Cordova CD, Lindsay H, Schmidt TS, Brugiroux S, Periaswamy B, Bauer R, Sturm A, Schreiber F, et al. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe. 2013;14:641–651. doi: 10.1016/j.chom.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, Fischbach MA, Sonnenburg JL. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis JF, Gros P. Genetic analysis of resistance to infections in mice: A/J meets C57BL/6J. Curr Top Microbiol Immunol. 2008;321:27–57. doi: 10.1007/978-3-540-75203-5_2. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli : identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect Immun. 2004a;72:1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K 12 (MG1655) in the mouse intestine. Infect Immun. 2004b;72:1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina PE. Endocrine Physiology, second edn. The McGraw Hill Companies Inc.; 2006. [Google Scholar]
- Moreira CG, Weinshenker D, Sperandio V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect Immun. 2010;78:914–926. doi: 10.1128/IAI.01038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushin R, Dubos R. Colonization of the mouse intestine with Escherichia coli. J Exp Med. 1965;122:745–757. doi: 10.1084/jem.122.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology. 2009;155:521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- Nakano M, Takahashi A, Sakai Y, Nakaya Y. Modulation of pathogenicity with norepinephrine related to the type III secretion system of Vibrio parahaemolyticus. J Infect Dis. 2007;195:1353–1360. doi: 10.1086/513275. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoroge J, Sperandio V. Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect Immun. 2012;80:688–703. doi: 10.1128/IAI.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoroge JW, Gruber C, Sperandio V. The interacting Cra and KdpE regulators are involved in the expression of multiple virulence factors in enterohemorrhagic Escherichia coli. J Bacteriol. 2013;195:2499–2508. doi: 10.1128/JB.02252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. MBio. 2012;3:e00280–e00212. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavia AT, Shipman LD, Wells JG, Puhr ND, Smith JD, McKinley TW, Tauxe RV. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J Infect Dis. 1990;161:255–260. doi: 10.1093/infdis/161.2.255. [DOI] [PubMed] [Google Scholar]
- Pepin J, Saheb N, Coulombe MA, Alary ME, Corriveau MP, Authier S, Leblanc M, Rivard G, Bettez M, Primeau V, et al. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin Infect Dis. 2005;41:1254–1260. doi: 10.1086/496986. [DOI] [PubMed] [Google Scholar]
- Pifer R, Sperandio V. The Interplay between the Microbiota and Enterohemorrhagic Escherichia coli. Microbiol Spectr. 2014;2(5) doi: 10.1128/microbiolspec.EHEC-0015-2013. [DOI] [PubMed] [Google Scholar]
- Pultz NJ, Hoskins LC, Donskey CJ. Vancomycin-resistant Enterococci may obtain nutritional support by scavenging carbohydrate fragments generated during mucin degradation by the anaerobic microbiota of the colon. Microb Drug Resist. 2006;12:63–67. doi: 10.1089/mdr.2006.12.63. [DOI] [PubMed] [Google Scholar]
- Purves D, Fitzpatrick D, Williams SM, McNamara JO, Augustine GJ, Katz LC, LaMantia A. Neuroscience, second edn. Sinauer Associates, Inc.; 2001. [Google Scholar]
- Ramseier TM, Bledig S, Michotey V, Feghali R, Saier MH., Jr The global regulatory protein FruR modulates the direction of carbon flow in Escherichia coli. Mol Microbiol. 1995;16:1157–1169. doi: 10.1111/j.1365-2958.1995.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Ramseier TM, Negre D, Cortay JC, Scarabel M, Cozzone AJ, Saier MH., Jr In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, pts and icd operons of Escherichia coli and Salmonella typhimurium. J Mol Biol. 1993;234:28–44. doi: 10.1006/jmbi.1993.1561. [DOI] [PubMed] [Google Scholar]
- Rasko DA, Moreira CG, Li de R, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading NC, Rasko DA, Torres AG, Sperandio V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci U S A. 2009;106:5889–5894. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, Louis P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. Isme J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, et al. The composition of the gut microbiota throughout life, with emphasis on early life. Microbial ecology in health and disease. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Ramseier TM. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996a;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Ramseier TM. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996b;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinner SA, Mokszycki ME, Adediran J, Leatham-Jensen M, Conway T, Cohen PS. Escherichia coli EDL933 requires gluconeogenic nutrients to successfully colonize the intestines of streptomycin-treated mice precolonized with E. coli Nissle 1917. Infect Immun. 2015;83:1983–1991. doi: 10.1128/IAI.02943-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014;20:719–730. doi: 10.1016/j.cmet.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Song Y, Garg S, Girotra M, Maddox C, von Rosenvinge EC, Dutta A, Dutta S, Fricke WF. Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent Clostridium difficile infection. PLoS One. 2013;8:e81330. doi: 10.1371/journal.pone.0081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Freitag N. Cell regulation. Curr Opin Microbiol. 2012;15:115–117. doi: 10.1016/j.mib.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003a;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A. 2003b;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Yen H, Tobe T. LeuO enhances butyrate-induced virulence expression through a positive regulatory loop in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2014;93:1302–1313. doi: 10.1111/mmi.12737. [DOI] [PubMed] [Google Scholar]
- Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, J ZL, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe T, Nakanishi N, Sugimoto N. Activation of motility by sensing short-chain fatty acids via two steps in a flagellar gene regulatory cascade in enterohemorrhagic Escherichia coli. Infect Immun. 2011;79:1016–1024. doi: 10.1128/IAI.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- Walters M, Sircili MP, Sperandio V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668–5681. doi: 10.1128/JB.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick LM, Qi W, Lacher DW, Whittam TS. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol. 2005;187:1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. Altering host resistance to infections through microbial transplantation. PLoS One. 2011;6:e26988. doi: 10.1371/journal.pone.0026988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrun SD, Melton-Celsa AR, Smith MA, Gilbreath JJ, Merrell DS, O'Brien AD. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc Natl Acad Sci U S A. 2013;110:E2126–E2133. doi: 10.1073/pnas.1222014110. [DOI] [PMC free article] [PubMed] [Google Scholar]