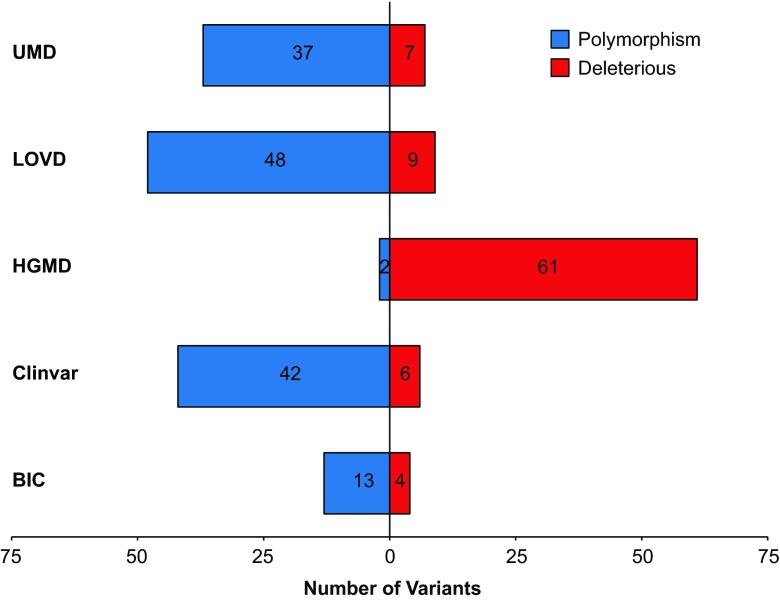
Fig. 3.
Analysis of the LSDB clinical classifications (benign or pathogenic) of variants considered “VUS” by the criteria outlined in the “Methods” section. Sixty-three of the 124 unique variants shared across the BIC, ClinVar, HGMD (paid version), LOVD, and UMD databases met these criteria. To determine how LSDBs vary in their treatment of these “challenging to classify” variants, we compared the clinical classifications of each “VUS” in all five databases. Variants with conflicting entries of VUS and either pathogenic or benign in the same database are categorized as either pathogenic or benign in the figure (see Table 1). Variants with conflicting classifications of both pathogenic and benign in the same database were excluded from this figure