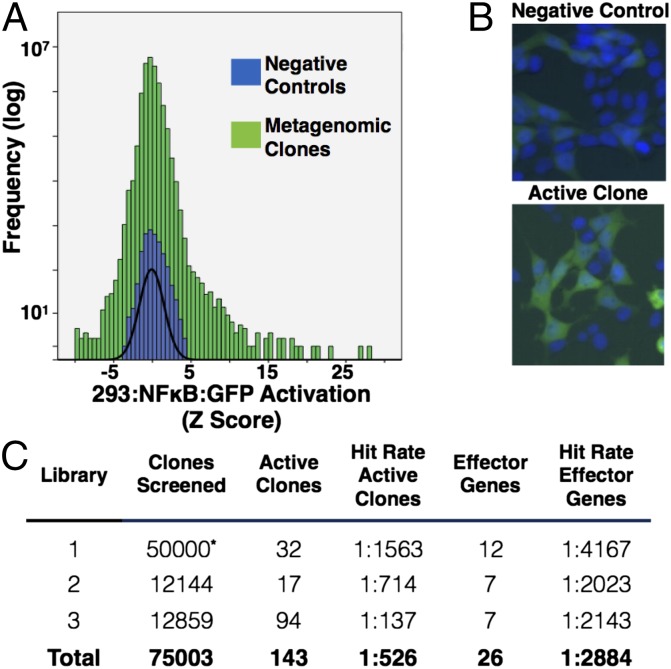
Fig. 2.
Screen results. (A) Histogram of HEK293:NF-κB:GFP reporter activation Z scores for all 75,003 metagenomic clones screened in this study. The percentage of activated cells was determined on a per-well basis and normalized to negative control wells on the assay plate to give a Z score. The Z-score distribution from metagenomic clones demonstrates a positive skew toward increased GFP activation compared with negative control wells (normal curve, black line). (B) Images of a representative negative control well and a well from an active metagenomic clone. Nuclei appear blue (Hoechst 33442 stain). HEK293:NF-κB:GFP cells expressing GFP appear green. (C) Table of total metagenomic clones screened and hit rates for each library. Active clones have reproducible HEK293:NF-κB:GFP cell activation in a secondary assay. Effector genes were identified by transposon mutagenesis of unique cosmids recovered from the active hits. All hit rates are expressed relative to total metagenomic clones screened. *Library 1 was not robotically arrayed into a microplate but mechanically dispensed at a dilution of 0–3 metagenomic clones per well. Total number of clones screened in library 1 is therefore an estimate based on the number of wells screened and average number of clones per well.