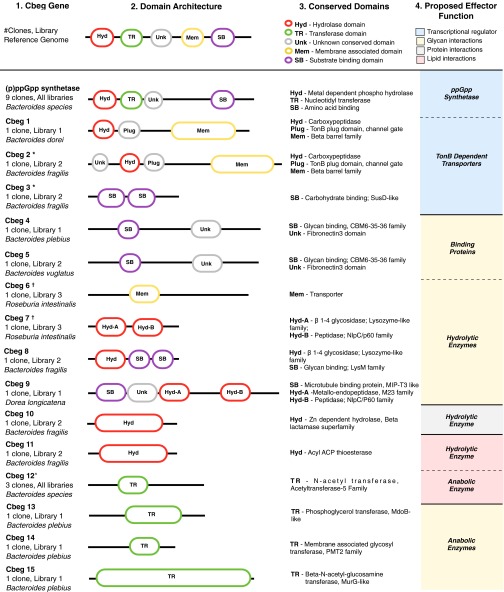
Fig. 4.
Summary of commensal bacterial effector genes (Cbegs). Each effector gene was queried by BLASTn against the NCBI nr dataset to determine the reference genome from which the most closely related sequence arises (column 1) and to predict the domain architecture and makeup of each Cbeg protein (columns 2 and 3, respectively). The metagenomic library from which each Cbeg was recovered is shown in column 1. In column 4, we have grouped Cbegs based on their general predicted functions. *Cbeg2 and Cbeg3 are found on the same unique cosmid. †Cbeg6 and Cbeg7 are found on the same unique cosmid. ^Cbeg12 is found at 98% identity in three unique cosmids, each in a different patient library (Cbeg12-1, Cbeg12-2, Cbeg12-3).