Fig. 6.
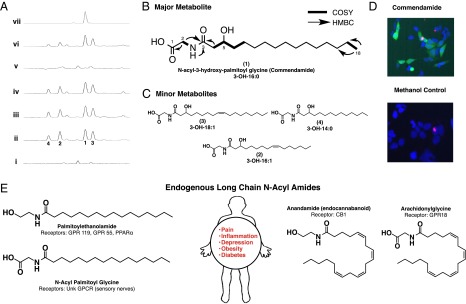
Characterization of commendamide. (A) Electrospray ionization (ESI)–mass spectroscopy (MS) traces of culture broth extracts from E. coli transformed with an empty pJWC1 cosmid vector (i), cosmid Cbeg12-1 (ii), cosmid Cbeg12-2 (iii), cosmid Cbeg12-3 (iv), cosmid Cbeg12-1 with a transposon insertion in the Cbeg12-1 gene (v), Cbeg12-1 subcloned into pJWC1 (vi), and synthetic commendamide (vii). (B) Key NMR correlations used to define the structure of commendamide (1), the major clone-specific peak found in cultures of E. coli transformed with Cbeg12-1, are shown. (C) The structures for three minor clone-specific metabolites related to commendamide (compounds 2–4) were also determined using NMR and MS data. (D) Purified commendamide activates the HEK293:NFκB GFP reporter assay. (E) Endogenous long-chain N-acyl-amides that are structurally related to commendamide are reported to function as agonists for numerous receptors, including many GPCRs. Such signaling systems have been therapeutically targeted for the treatment of pain, inflammation, depression, obesity, and diabetes.
