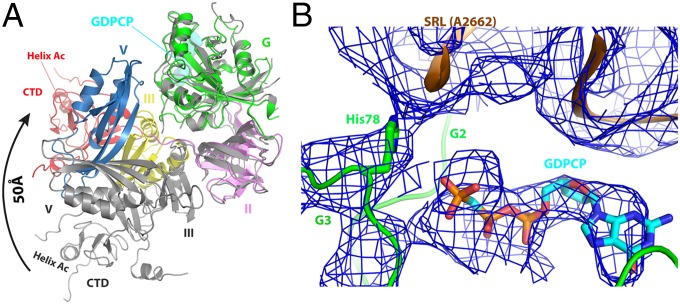
Fig. 3.
Structure of ribosome-bound BipA in GTP form. (A) Conformational changes and domain rearrangements in BipA upon ribosome binding. Ribosome-bound BipA (colored as in Fig. 1) and isolated BipA (gray) are aligned based on G domain. (B) BipA G domain switch I (G2 motif) and switch II (G3 motif) region interactions with GDPCP and 50S subunit SRL. The A2662 residue of SRL and the proposed catalytic His78 residue of BipA G3 motif are highlighted. The corresponding cryo-EM reconstitution density map is shown in blue mesh.