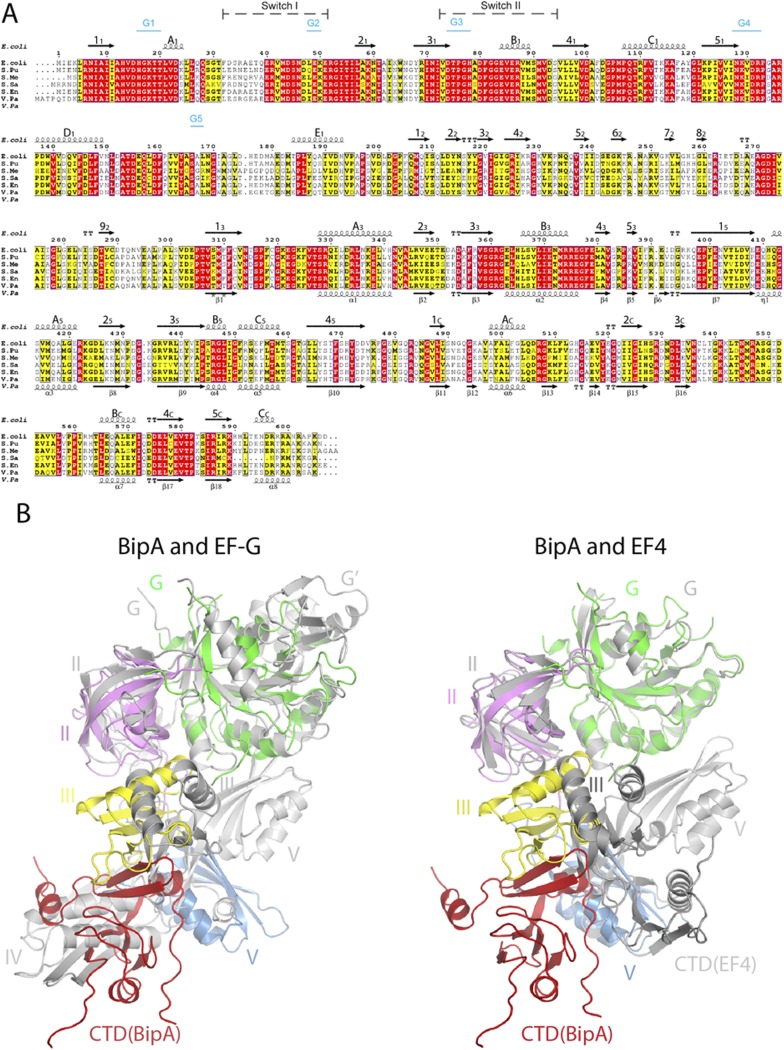
Fig. S1.
BipA sequence alignment and structure comparison with EF-G and EF4. (A) Sequence alignment of BipA from various bacteria (E. coli, Escherichia coli; S. Pu, Pseudomonas putida; S. Me, Sinorhizobium meliloti; S. Sa, Suaeda salsa; S. En, Salmonella enterica subspecies Enterica typhimunium; V. Pa, Vibrio parahaemolyticus) was performed by using ClustalW. Conserved and semiconserved residues are highlighted in red and yellow, respectively. Secondary structure elements of full-length E. coli (shown above the sequence alignment) and C-terminal half of Vibrio parahaemolyticus (V. Pa) BipA proteins (shown below the sequence) are derived from the apo form BipA structure presented in this work, and the structure deposited in PDB (3E3X). α-Helices (springs) and β-sheets (arrows) are labeled. Switch regions and G-motifs are indicated as well. (B) Comparison of the overall structure of BipA with EF-G and EF4. BipA structure in apo form is superimposed (by aligning the G domains) with EF-G (PDB ID code: 2BMO) and EF4 (PDB ID code: 3CB4). BipA domains are colored as in Fig. 1. EF-G and EF4 are shown in gray cartoon.