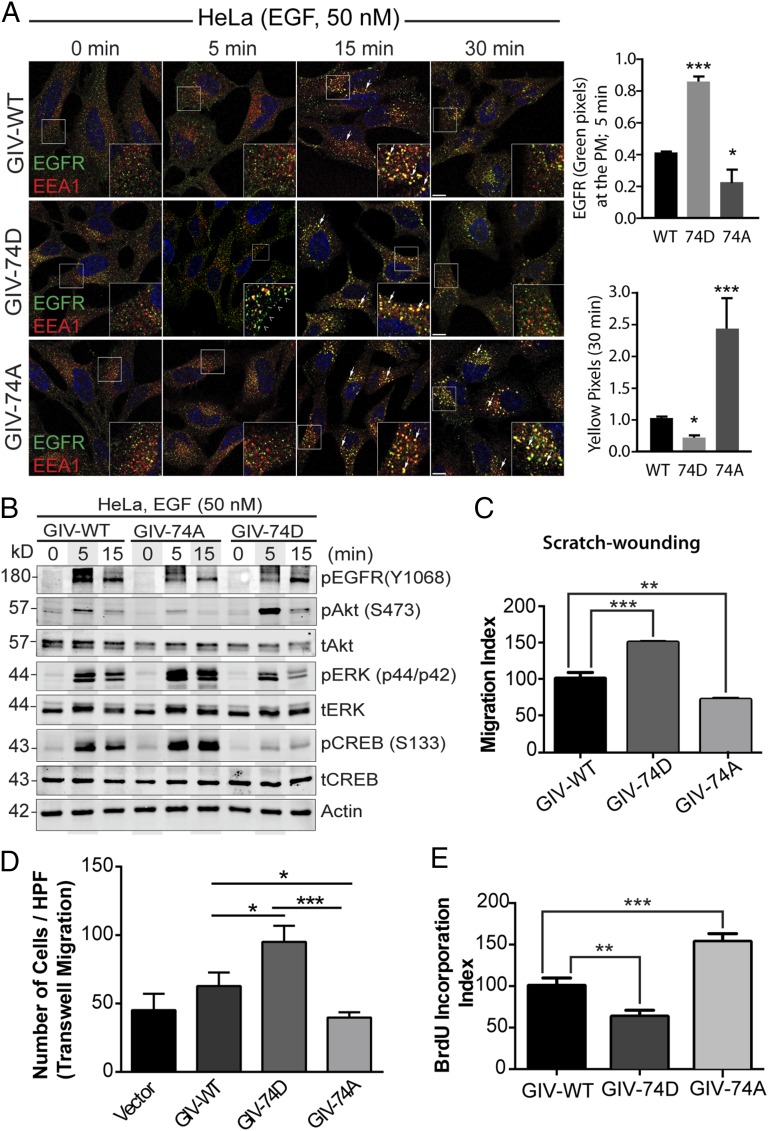
Fig. 2.
Phosphorylation of GIV at S1674 influences EGFR trafficking, signaling, and preference for migration versus proliferation of HeLa cells. (A) Serum starved (0.2% FBS, overnight) GIV-depleted HeLa cells stably expressing GIV-WT, 74D, and 74A (Fig. S2A) were stimulated with 50 nM EGF before fixation. Fixed cells were stained for total EGFR (green), the early endosome marker EEA1 (red), and DAPI (nucleus; blue) and examined by confocal microscopy. (Insets) Enlargement (3×) of boxed regions. (Scale bars, 10 μm.) Phosphomimetic 74D mutant GIV enhances the duration EGFR spends at the PM (arrowheads, green pixels at 5 min), whereas the nonphosphorylatable 74A mutant GIV prolongs the duration EGFR spends on early endosomes (arrow, yellow pixels at 30 min). Bar graphs on the Right show quantification of pixels in each set of cells at the indicated time points. (B) Serum starved, GIV-depleted (by shRNA) HeLa cell lines stably expressing shRNA-resistant GIV-WT, 74D, and 74A were stimulated with 50 nM EGF before lysis. Equal aliquots of whole-cell lysates were analyzed for phospho(pY1068)EGFR, total(t) and phospho(p)-Akt, -ERK, and -CREB and actin by immunoblotting. (C) HeLa cells in B were subjected to scratch wounding and examined immediately (0 h) and after 24 h by light microscopy (Fig. S2C). The migration index (y axis) was calculated as described in Experimental Procedures, normalized to cells expressing GIV-WT (set at 100%) and plotted as a bar graph. (D) HeLa cells in B were analyzed for chemotaxis toward EGF using a Transwell migration assay (Fig. S2D). Cells were allowed to migrate for 6 h, fixed, and stained with Toluidine Blue. The number of migrating cells was averaged from 20 field-of-view images per experiment. Data are presented as mean ± SEM; n = 3. HPF, high power field. (E) HeLa cells in B were grown in low serum [2% (vol/vol) FBS] overnight, incubated in BrdU for 30 min, fixed [3% (wt/vol) paraformaldehyde], stained for BrdU, and analyzed by confocal microscopy (Fig. S2E). Bar graph shows percentage of cells with BrdU uptake (y axis). Data are presented as mean ± SEM; n = 3.