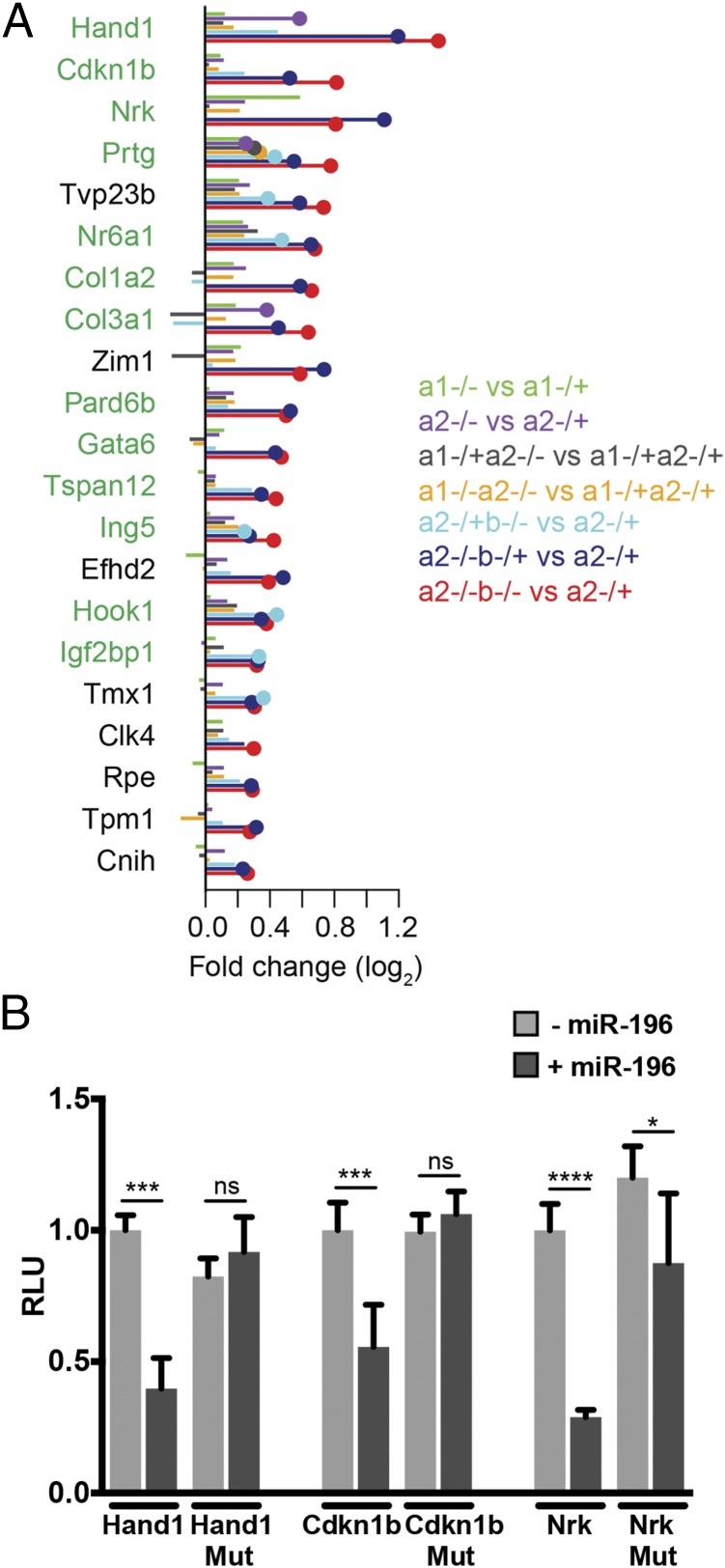
Fig. 5.
Identification of additional putative direct (non-Hox) miR-196 targets. (A) List of the most highly up-regulated genes and their associated fold changes in seven genotype comparisons that either (i) contain a conserved miR-196 binding site or (ii) are predicted to respond strongly to the miRNA (i.e., have a context+ score ≤ −0.2). Genes with one or more conserved miR-196 target binding sites are indicated in green. (B) In vitro luciferase analysis confirms sequence-specific regulation of three experimentally supported target genes of miR-196. Renilla luciferase intensity values have been normalized to their respective Firefly values (RLU). Controls (WT 3′ UTR construct without miR-196b) were set to 1. MUT, mutated 3′ UTR construct destroying miR-196 binding site. Error bars represent SD. P values, Student t test: *P < 0.05, ***P < 0.0005, and ****P < 0.0001.