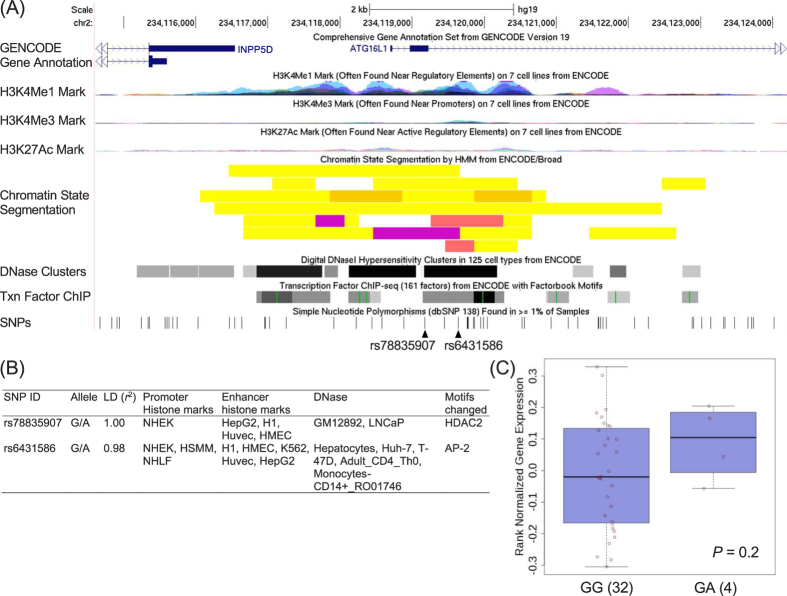
Figure 2. Summary of the functional analyses for the linkage disequilibrium (LD) block containing ATG16L1 rs78835907.
(A) Expanded view of the ENCODE data for the LD block containing the ATG16L1 rs78835907. The H3K4Me1, H3K4Me3, and H3K27Ac tracks show the genome-wide levels of enrichment of the mono-methylation of lysine 4, tri-methylation of lysine 4, and acetylation of lysine 27 of the H3 histone protein, as determined by the ChIP-seq assays. These levels are thought to be associated with promoter and enhancer regions. Chromatin State Segmentation track displays chromatin state segmentations by integrating ChIP-seq data using a Hidden Markov Model for H1 embryonic stem cells, HepG2 hepatocellular carcinoma cells, HUVEC umbilical vein endothelial cells, HMEC mammary epithelial cells, HSMM, skeletal muscle myoblasts, NHEK epidermal keratinocytes, and NHLF lung fibroblasts. The chromatin state regions predicted for promoters and enhancers are highlighted. DNase clusters track shows DNase hypersensitivity areas. Tnx Factor track shows regions of transcription factor binding of DNA, as assayed by ChIP-seq experiments. (B) Regulatory annotation of variants within the LD block containing ATG16L1 rs78835907. In the LD block with the lead SNP rs78835907, ENCODE data showed evidence of promoter and enhancer elements coinciding with the variants in many different cell types. In addition, HDAC2 and AP-2 motifs are predicted to be affected. (C) Expression quantitative trait locus association between rs78835907 genotype and ATG16L1 expression in prostate tissues (GTEx data set). Numbers in parentheses indicate the number of cases.