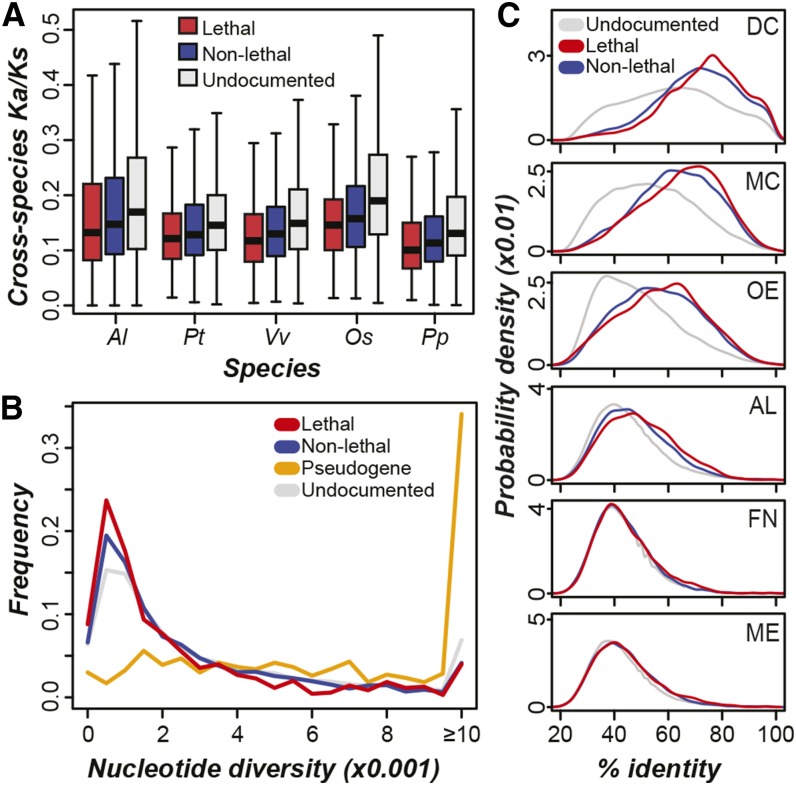
Figure 4.
Evolutionary Rate and Cross-Species Protein Conservation of A. thaliana Phenotype Genes.
(A) Ratios of nonsynonymous substitutions (Ka) to synonymous substitutions (Ks) between A. thaliana genes and homologs in the same OrthoMCL cluster from Arabidopsis lyrata (Al; KST of lethal versus nonlethal, P < 0.02), Populus trichocarpa (Pt; P < 0.01), Vitis vinifera (Vv; P < 0.01), O. sativa (Os; P < 0.02), and Physcomitrella patens (Pp; P < 0.04). Lower Ka/Ks values are indicative of stronger negative selection pressure.
(B) Distributions of nucleotide diversity for lethal, nonlethal, pseudo-, and undocumented genes. Higher nucleotide diversity values indicate higher degree of sequence polymorphism between A. thaliana accessions.
(C) Probability density distributions of median percentage of identity of lethal, nonlethal, and undocumented genes to top BLASTP matches in dicotyledonous plants (DC; lethal versus nonlethal KST, P < 2e-6), monocotyledonous plants (MC; P < 7e-4), other embryophytic plants (OE; P = 0.05), algae (AL; P < 7e-6), fungi (FN; P = 0.07), and metazoans (ME; P = 0.25).