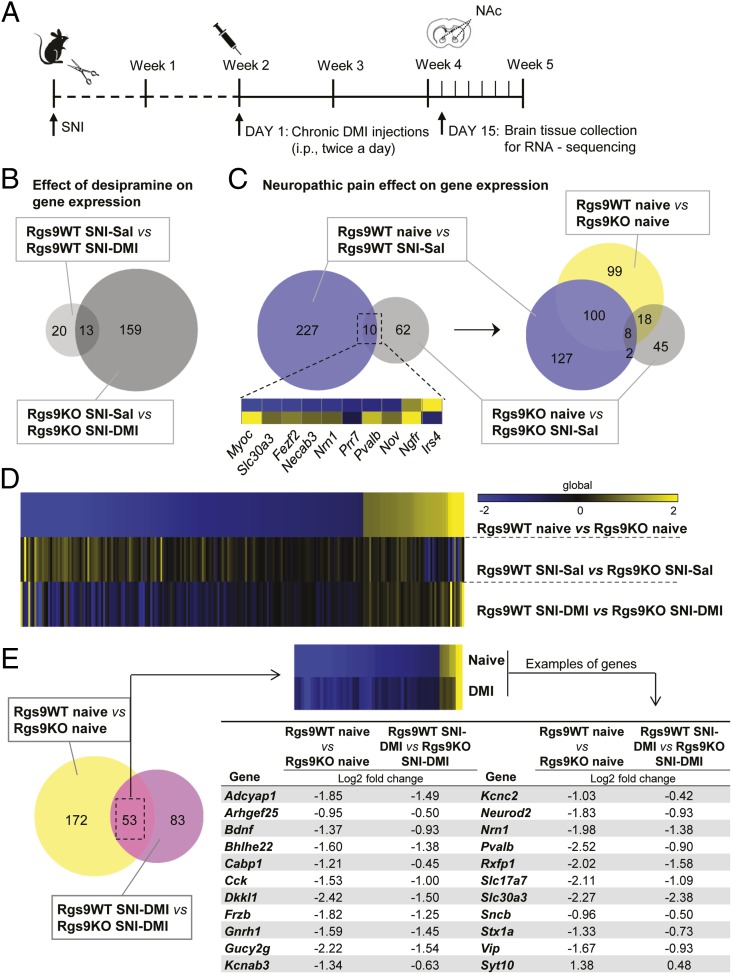
Fig. 5.
NAc-specific RNA-seq profiling of Rgs9 knockout in naive and neuropathic pain mice, treated either with saline or DMI. (A) Timeline of the RNA-seq experiments. Briefly, NAc tissue was collected at the time point when only the mutant mice responded to the chronic DMI treatment. (B) Venn diagram depiction of the effect of drug treatment on gene expression. As expected, gene expression was slightly affected in the NAc of Rgs9WT SNI–DMI mice compared with saline-treated controls, because tissue collection was performed at the time point at which only the mutant mice developed antiallodynic response. (C) Neuropathic pain effect on gene expression in drug-naive mice. Venn diagram between the groups of naive and neuropathic pain-treated Rgs9WT and Rgs9KO mice suggests that the effect of SNI on gene expression was less potent in the NAc of mutant mice, whereas SNI affected different genes in the wild-type vs. mutant mice. The small embedded heatmap shows that even most of the 10 overlapping genes change to opposite direction. When differentially expressed genes between naive Rgs9WT and Rgs9KO mice were added to the Venn diagram, it was revealed that half of the adaptations induced after SNI surgery in wild-type mice are already present in the NAc of the naive mutant mice, possibly explaining the small effect in the Rgs9KO NAc after SNI. (D) Heatmap analysis reveals that the gene regulation pattern in the NAc of Rgs9KO naive mice is very similar to the gene regulation induced by DMI treatment, suggesting that NAc-specific gene adaptations in the absence of Rgs9 promote DMI’s actions. (E, Left) Venn diagram of the genes from naive and SNI–DMI groups of Rgs9WT and Rgs9KO mice reveals an overlap of 53 genes with very similar regulation, as indicated by the embedded heatmap. (E, Right) The table lists some of these genes with the corresponding fold changes. Importantly, the direction of the change is similar between the groups of naive and SNI–DMI-treated mice, suggesting that this adaptive gene regulation in the Rgs9KO naive mice facilitates DMI’s antiallodynic effect.