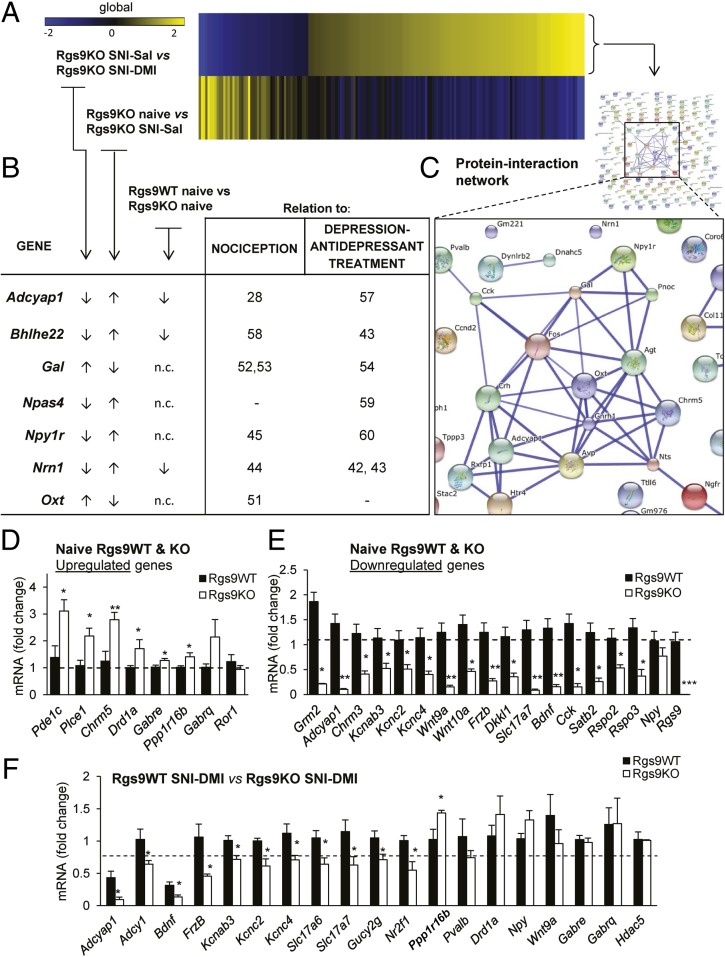
Fig. 6.
Rgs9 gene knockout promotes adaptive changes that resemble DMI’s therapeutic effect. (A) Heatmap of the genes regulated by chronic DMI in the NAc of Rgs9KO SNI group reveals that chronic antidepressant treatment almost completely reversed the adaptive changes in the group of mutant mice under neuropathic pain, suggesting that ablation of Rgs9 promotes adaptive changes that facilitate DMI’s actions. (B) Table depicting fold changes for the genes between the aforementioned group comparison as well as the naive group of mice. Importantly, the direction of the change for many genes is similar between the first and the third group comparisons shown in the table, suggesting once again that adaptive changes in the NAc of Rgs9KO naive mice are beneficial for the action of DMI. (C) Moreover, by using the STRING database for protein–protein interactions, a highly interconnected network of gene products from the Rgs9KO SNI–DMI compared with Rgs9KO SNI–Sal group was revealed. (D and E) To validate the RNA-seq results, several genes from the groups of naive Rgs9WT vs. Rgs9KO and Rgs9WT SNI–DMI vs. Rgs9KO SNI–DMI mice were selected to be tested with qPCR. (D) Up-regulated genes. (E) Down-regulated genes. *P < 0.05; **P < 0.01; ***P < 0.001 (t test, unpaired two-tailed). (F) Similarly, 18 of 22 selected genes (19 genes shown here) from the group of mice that responded to DMI treatment were confirmed by qPCR (data expressed as log2 fold change). *P < 0.05 (t test, unpaired two-tailed). N.c., no change.