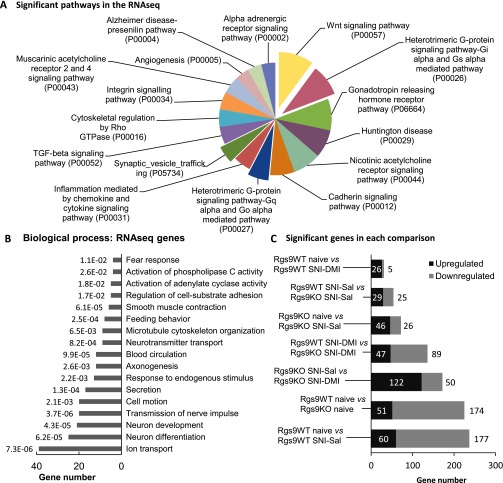
Fig. S3.
GO and experimental groups of the RNA-seq study. (A) To gain an overview of the pathways affected by our genetic or pharmacological manipulations, GO analysis was performed for all of the significantly altered genes identified from the RNA-seq analysis, regardless of experimental group. The analysis revealed changes in the expression of genes that play a role in antidepressant drug responses or mood or pain disorders, including Wingless-related integration site signaling pathway (Wnt) signaling, alpha-adrenergic receptor, and heterotrimeric G-protein–signaling pathways. Only pathways that reached statistical significance are shown. (B) The same approach was followed to unravel which biological processes were most represented in the RNA-seq results. The enriched biological processes, along with the assigned P value and the number of genes for every GO term, are shown on the graph. (C) Summary of the main group comparisons of the study, including the total number of significant up- and down-regulated genes within each group. As expected, the least change was observed in the comparison between Rgs9WT SNI–Sal and Rgs9WT SNI–DMI groups, because at this time point, wild-type mice fail to show an antiallodynic response. On the contrary, most gene expression changes were observed between naive Rgs9WT and naive Rgs9KO mice and between naive Rgs9WT and Rgs9WT SNI–Sal groups.