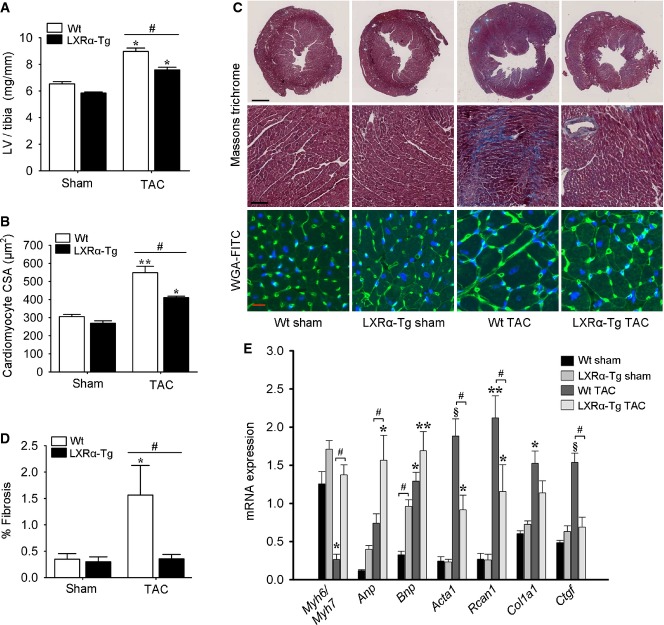
Figure 2. LXRα overexpression attenuates TAC-induced cardiac hypertrophy and fibrosis after 5 weeks.
- LV/tibia ratios in sham- and TAC-operated Wt and LXRα-Tg mice; n = 21 Wt sham, n = 24 LXRα-Tg sham, n = 24 Wt TAC, n = 26 LXRα-Tg TAC. *P < 0.00001 versus respective sham, #P = 0.00004.
- Quantification of cardiomyocyte cross-sectional area from WGA-FITC-stained histological sections; n = 5 Wt sham, n = 4 LXRα-Tg sham, n = 5 Wt TAC, n = 3 LXRα-Tg TAC. **P = 0.00001 versus Wt sham, *P = 0.01 versus LXRα-Tg sham, #P = 0.01.
- Representative LV sections stained with WGA-FITC and Masson’s trichrome to assess cardiomyocyte hypertrophy and fibrosis, respectively; scale bars = 1 mm, 100 μm, 10 μm, from top to bottom.
- Whole area percentage of fibrosis in Masson’s trichrome-stained sections; n = 8 per group, except n = 7 in Wt TAC group. *P = 0.02 versus Wt sham, #P = 0.02.
- Measurement of mRNA levels with RT–PCR to assess induction of fetal gene program, as well as hypertrophy (Rcan1) and fibrosis (Col1a1, Ctgf); n = 8 per group, except n = 7 in Wt TAC group. Myh6/Myh7: *P = 0.0001 versus Wt sham, #P = 0.00001; Anp: *P = 0.0005 versus LXRα-Tg sham, #P = 0.02; Bnp: *P = 0.0006 versus Wt sham, **P = 0.009 versus LXRα-Tg sham, #P = 0.03; Acta1: §P < 0.00001 versus Wt sham, *P = 0.01 versus LXRα-Tg sham, #P = 0.001; Rcan1: **P = 0.00004 versus Wt sham, *P = 0.05 versus LXRα-Tg sham, #P = 0.04; Col1a1: *P = 0.00003 versus Wt sham; Ctgf: §P < 0.00001 versus Wt sham, #P = 0.00001.
Data information: Data are means ± SEM; one-way ANOVA with Bonferroni’s multiple comparison test was used to compare groups.