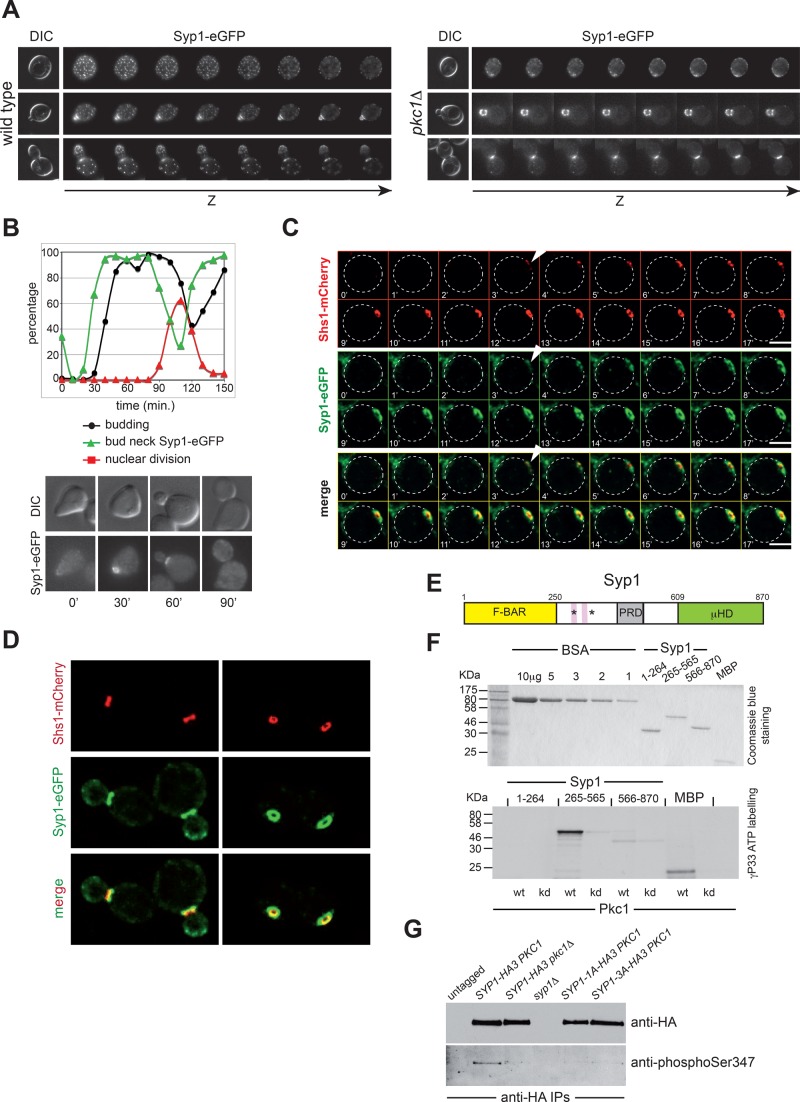
FIGURE 5:
Pkc1 phosphorylates Syp1 and controls its bud neck localization. (A) Z-stack serial images of wild-type and pkc1Δ cells expressing Syp1-eGFP at different cell cycle stages (top: G1, unbudded; middle: S phase, small budded; bottom: mitosis, large budded). Spacing between sequential planes is 0.3 μm. (B) Wild-type cells expressing Syp1-eGFP were arrested in G1 by α-factor and released in fresh YEPD medium at 25°C. At the indicated time points, cells were collected for FACS analysis of DNA contents (not shown) and kinetics of budding, nuclear division, and Syp1-eGFP localization. (C) Wild-type cells expressing Syp1-eGFP and Shs1-mCherry were filmed at room temperature with 1-min time lapse. Z-stacks (31 planes at 0.2-μm spacing) were maximum projected and deconvolved with Huygens. Arrowheads indicate the appearance of septin and Syp1 rings. (D) Images of wild- type cells expressing Syp1-eGFP and Shs1-mCherry show that Syp1 rings are larger in diameter than Shs1 rings and located on mother side of the bud neck. (E) Schematic representation of the Syp1 protein showing the N-terminal F-BAR domain (yellow), the middle, unstructured region (white) containing the proline-rich domain (gray), and the muniscin-homology domain (green) at the C-terminus. Pink boxes indicate the two phospholipid-binding motifs, and asterisks mark the position of the serines phosphorylated by Pkc1. (F) The three domains of Syp1 were purified from bacteria (purified proteins are shown on top after Coomassie blue staining of an acrylamide gel) and subjected to in vitro phosphorylation by wild-type or kinase-dead (kd) Pkc1 immunoprecipitated from yeast cells ([γ-33P]ATP labeling, bottom). (G) Syp1-HA3 was immunoprecipitated from protein extracts obtained from cycling cultures of strains with the indicated genotypes and probed by Western blot with a phospho-specific antibody raised against phosphorylated Ser-347, as well as with anti-HA antibodies.