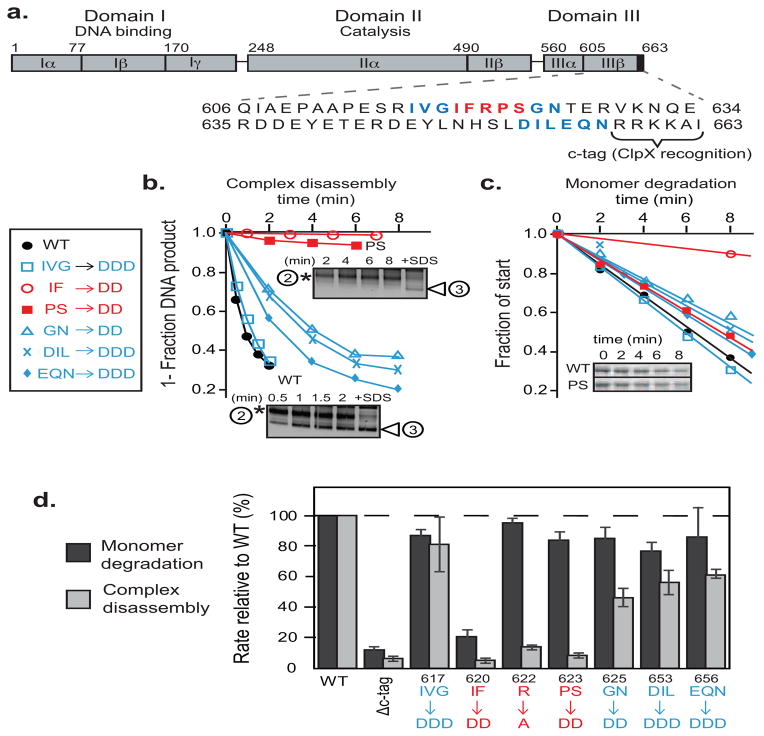
FIGURE 2. A sequence region Ile620-Ser624 forms a critical interaction between MuA complexes and ClpX.
A) MuA transposase is a ~75kDa protein comprised of three domains. Domain III contains the C-terminal ClpX pore-binding tag, “C-tag”, which is comprised of the last six residues. Sequences that were mutated in this study are in bolded red or blue.
B) ClpX-catalyzed disassembly of complexes that were assembled from wild-type MuA and MuA “aspartate” variants. Initial transpososome concentration was 100nM. Quantification of appearance of the lowermost DNA disassembly product (white arrow) over time. Two representative native agarose gels of wild-type MuA complexes and MuA(P623D, S624D) mutant complexes are shown. The “+SDS” lane shows the pattern of topoisomer migration upon complete disassembly.
C) Degradation of wild-type MuA and MuA “aspartate” variants by ClpXP protease at subsaturating substrate concentrations (1uM). Inset shows a representative SDS-PAGE gel of wild-type MuA and MuA(P623D, S624D) monomers.
D) Quantification of differences in degradation and disassembly rates of MuA variants whose indicated sequences were mutated to alanine or aspartic acid relative to wild-type MuA. Reactions were performed in triplicate. Error bars are the standard error of the mean.