FIGURE 5. Design and construction of SinMu chimeric transpososomes.
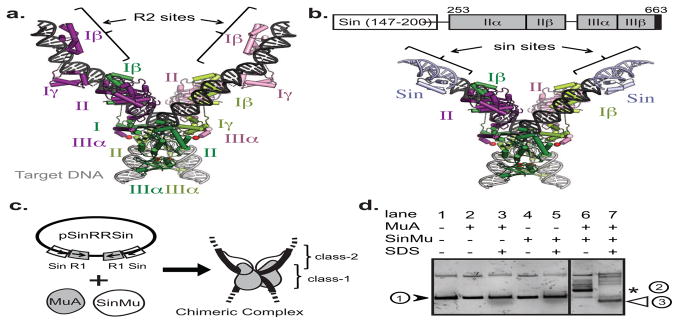
A) Crystal structure of STC transpososome on two “right-end” oligos (PDB 4FCY) with each subunit in a different color. Subdomains Iβ and Iγ are sequence-specific DNA-binding domains. Pink and purple subunits are bound to R2 sites; green ones to R1 sites. Target DNA is in grey.
B) Model of a chimeric transpososome containing SinMu protein and MuA on DNA fragments carrying the Mu-R1 sites and the Sin-R2 sites. Domain structure of the SinMu chimeric protein, which contains the DNA binding domain of Sin resolvase (147–200; from PDB 2r0q) followed by a linker and MuA domains II and III.
C) The altered- specificity plasmid substrate pSinRRSin has the MuA binding sites, L2 and R2, swapped out for Sin-specific DNA binding sites. This arrangement restricts SinMu protein to the class-2 structural subunits and MuA protein to the class-1 catalytic subunits.
D) Assembly of chimeric complexes on pSinRRSin plasmid requires the presence of both MuA and SinMu proteins. On a native agarose gel, lane 1 contains un-reacted pSinRRSin supercoiled plasmid (black arrow). The in vitro assembly reaction shows the band associated with assembled complexes (asterisk) in lane 6, but not in lanes 2 and 4 which each lack one species of protein. The characteristic pattern of recombined DNA disassembly products (white arrow) from a reaction identical to the one in lane 6 is seen after addition of SDS in lane 7.
