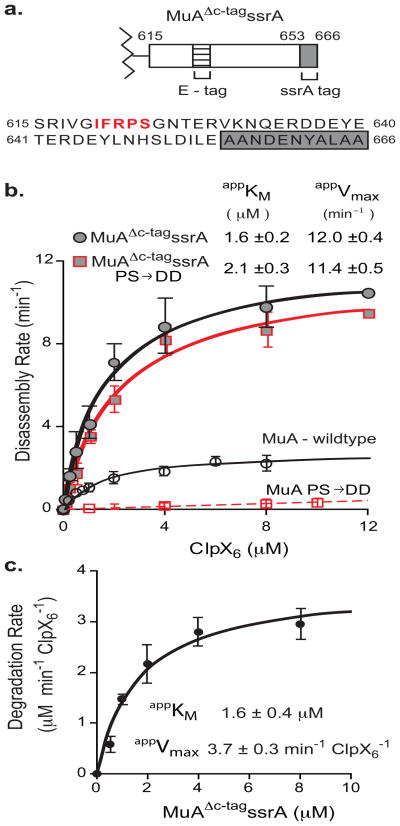
FIGURE 7. ClpX loses discrimination between monomers and transpososomes with a strong Ctag recognition signal.
A) Diagram of MuAΔc-tagssrA zoomed in on domain IIIβ and corresponding sequence below. E-tag is in bold red. ssrA tag is in grey box.
B) Half-maximal velocity determination for disassembly of wild-type, MuA (PS/DD), MuAΔc-tagssrA, and MuAΔc-tagssrA (PS/DD) complexes by ClpX. Data for wild-type and MuA (PS/DD) are superimposed from Figure 2D. Reactions were repeated four times. Error bars at each concentration point are standard deviation of the average. Errors of the appKM are standard deviation of the average of four fits.
C) Half-maximal velocity determination for degradation of MuAΔc-tagssrA monomers by ClpXP. ClpX6 was 0.3μM, ClpP14 was 0.8μM. Error bars at each concentration point are standard deviation of the average. Error of the appKM is standard deviation of the average of three fits.