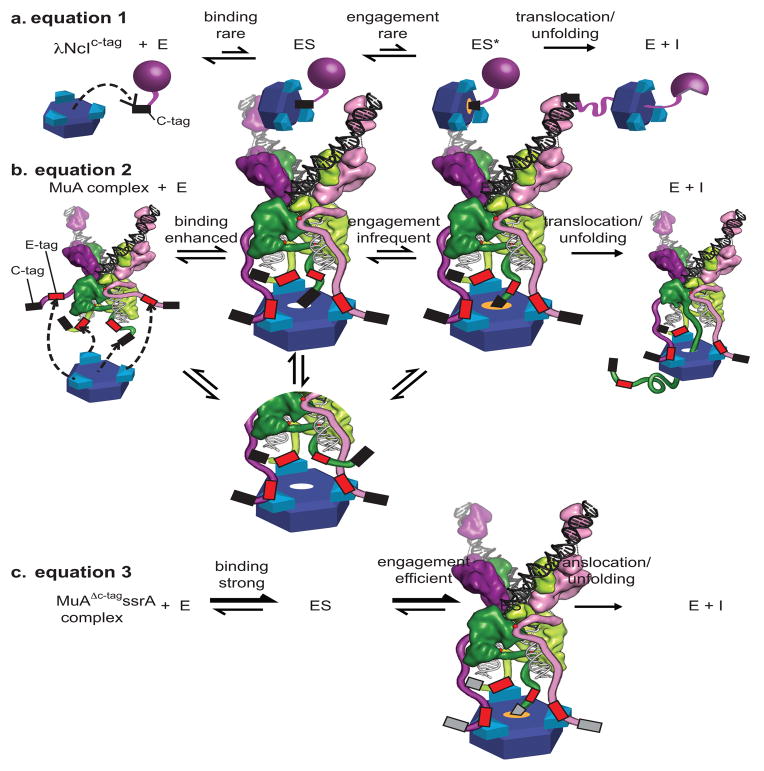
FIGURE 8. Kinetic Model of monomer versus transpososome recognition by ClpX.
A) λNcIc-tag monomer (purple sphere) binds weakly to the pore of ClpX (blue ring) and engagement is rare leading to weak apparent affinities. Engagement of the C-tag (depicted in an orange-glowing pore) leads to a transient enzyme-substrate complex (ES*). After engagement of the Ctag, ClpX processively translocates and unfolds the native protein.
B) Each N-domain dimer (turquoise) in ClpX can bind an E-tag in a wild-type MuA complex (colored as in Fig. 5, with cartooned extensions representing domain IIIβ, red boxes for E-tags and black boxes for C-tags). Binding of a C-tag to the pore of ClpX is enhanced because additional interactions between the E-tags and N-domains of ClpX raise the effective concentration of the C-tag near the pore by restricting the search volume needed for complex formation.
C) In the MuAΔc-tagssrA complex, binding of the ssrA tag to the pore of ClpX is strong. Engagement of the ssrA tag is strong and efficient and occurs similarly whether or not the E-tag is bound to the N-domains of ClpX.