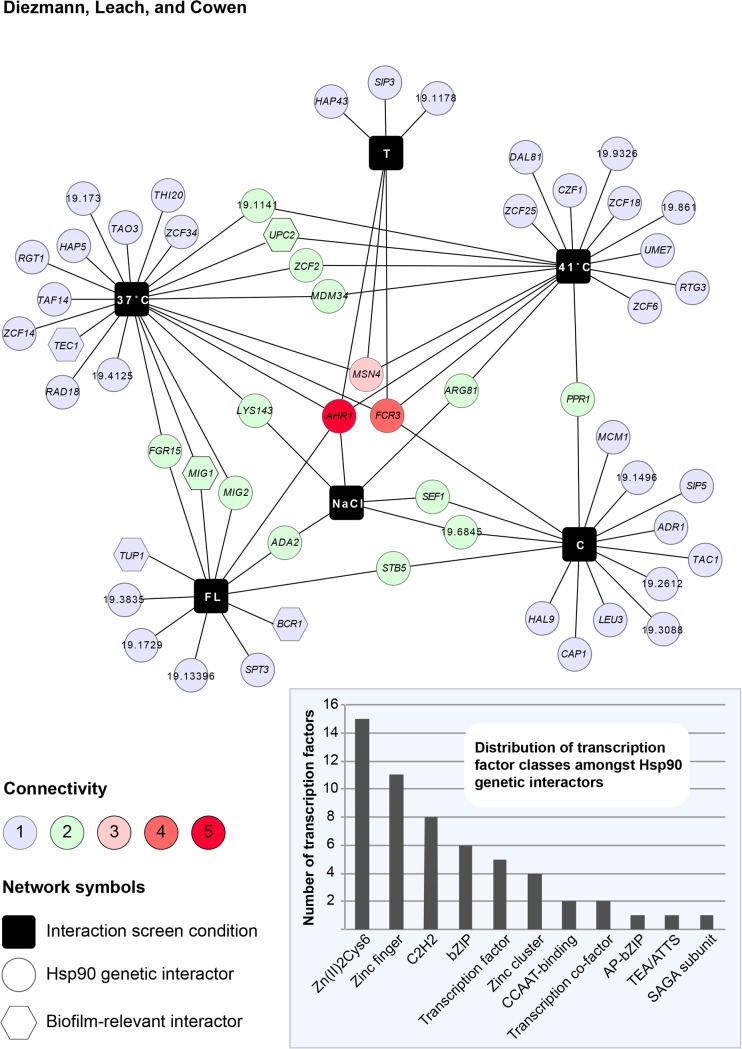
Fig 1. Hsp90 –transcription factors genetic interaction network.
Approximately one quarter of C. albicans Hsp90 genetic interactors are transcription factors. Of the 226 genetic interactors that were previously identified in a screen of 10% of the genome [21], 56 had GO category annotations for “transcription factor”. These were further analyzed with Cytoscape 2.8.1. and transformed into the network shown here. Interactors were mapped in six environmental conditions (black squares) representing normal growth (37°C), general stresses (41°C, NaCl), and specific stresses exerted by tunicamycin (T), caspofungin (C), and fluconazole (FL). Each interactor is connected by line(s) to the condition(s) it was originally identified in. An interaction that was scored multiple times has multiple connecting lines and is located in the center of the network. The degree of connectivity is also color-coded. The five transcription factors that were further here analyzed are represented as hexagons. Hsp90 interacts with different classes of transcription factors, with representatives of the Zn(II)2Cys6 and zinc finger families being the most common interactors (inset).