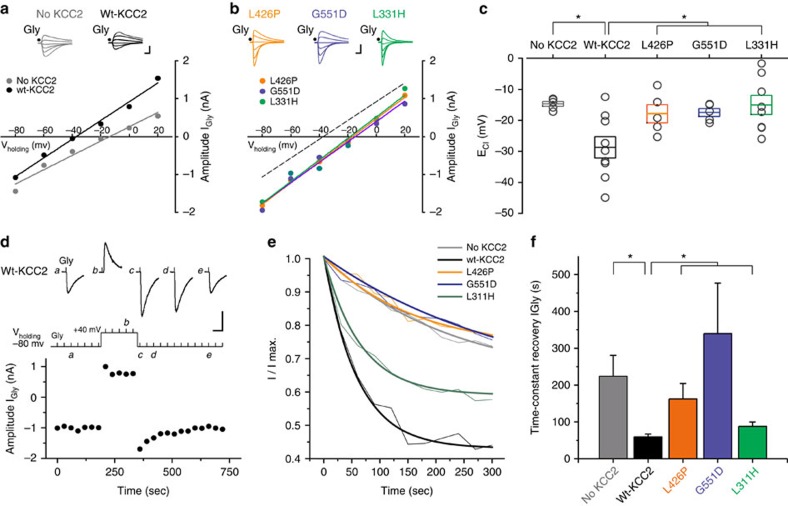
Figure 3. Electrophysiology of wild-type and mutant KCC2.
(a–c) KCC2 mutants exhibit a depolarized ECl. (a) Examples of I–V relationships in HEK293 cells transfected either with GlyR α2 alone (no KCC2), or GlyR α2 with a wild-type KCC2 construct. IGly amplitude was plotted against the holding potential and the intercept of the line of fit of this relation with the abscissa was taken as ECl. Superimposed traces show examples of glycine-evoked currents recorded from individual cells at different holding potentials (–80 to +20 mV). (b) Examples of I–V relationships in HEK293 cells co-transfected with GlyR α2 and mutant KCC2, as indicated. ECl in mutant KCC2 is depolarized in comparison to wild-type (fit of I–V relation indicated by dashed-line). Calibration bars for 3a and 3b: 1nA, 1sec. (c) Summary chart plotting the distribution of ECl for each cell group: no KCC2: –14.7±0.8 mV (n=5), GlyR-KCC2: –29±3.5 mV (n=9), G551D: –17.4±1.2 mV (n=5), L426P: –17.9±2.9 mV (n=5), L311H: –15±3.1 mV (n=9); *P=0.006, one-way analysis of variance (ANOVA). Grey symbols represent measurements from individual cells. Boxes represent the mean±standard error of the mean. GlyR α2: glycine receptor α2, Wt-KCC2: wild-type K+-Cl− co-transporter. (d–f) The rate of Cl− extrusion is impaired in KCC2 mutants. (d) IGly amplitude plotted against time in a HEK293 cell co-transfected with GlyRα2 and wild-type KCC2 and held at different membrane potentials as indicated in the top image. Vertical bars indicate the times of successive glycine applications. Example traces show glycine-evoked currents at times indicated by letters in italic. Calibration bars: 1 nA, 300 ms. (e) Rate of recovery of IGly amplitude after switching back the holding potential from +40 to −80 mV. Fine lines replace symbols and error bars are omitted for legibility. An exponential decay function was used to fit IGly amplitude ratio in each cell group. (f) Summary bar graph of the time-constant of IGly recovery in each group (no KCC2: 224±56.7 s, n=6; WT-KCC2: 59.5±7.7 s, n=13; L426P: 162.5±42.1 s, n=4; G551D: 340.1±137 s, n=4; L311H: 88.1±11.5 s, n=7). *Different from WT-KCC2 (P<0.01), ANOVA.