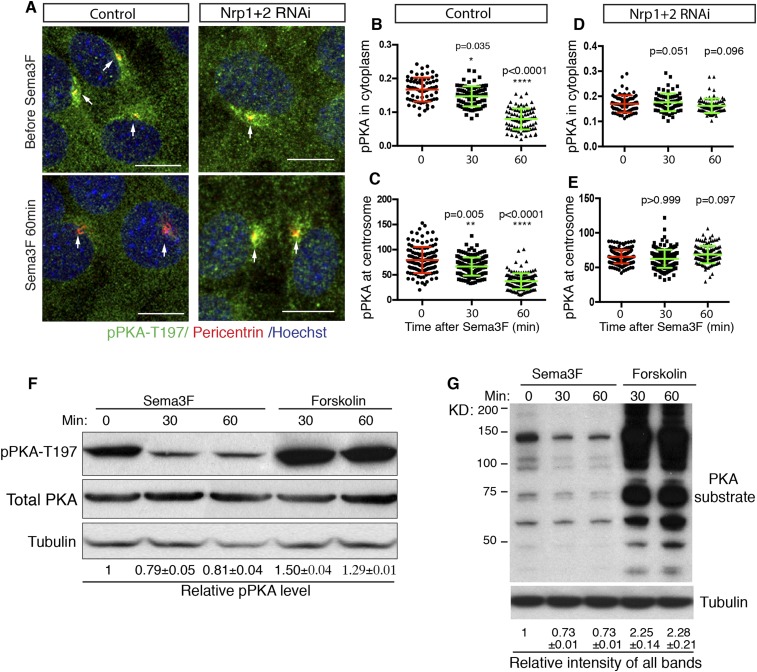
Figure 3. Sema3-Nrp signaling inhibits PKA activity.
(A) Immunofluorescence showing active PKA in NIH3T3 cells after Sema3F treatment. Cells were infected with lentiviruses expressing control shRNA or shRNA against Nrp1&2 for 72 hr. Active PKA is recognized by the antibody against phospho-PKA T197 (green). Scale bar, 10 μm. (B–E) Quantification of phospho-PKA level at the cytoplasm and centrosome. Data are shown as mean ± SD. Statistics: Kruskal–Wallis non-parametric One-Way ANOVA. (F) Western blot showing the phospho-PKA and total PKA levels in NIH3T3 cells treated with Sema3F or Forskolin for indicated time periods. Quantification of relative active PKA level (mean ± SEM) from three independent experiments was shown at the bottom of each blot. (G) Western blot showing the levels of PKA substrate phosphorylation in NIH3T3 cells treated with Sema3F or Forskolin for indicated time periods. Quantification of the relative intensity of all bands in each lane (mean ± SEM) from three independent experiments was shown at the bottom.