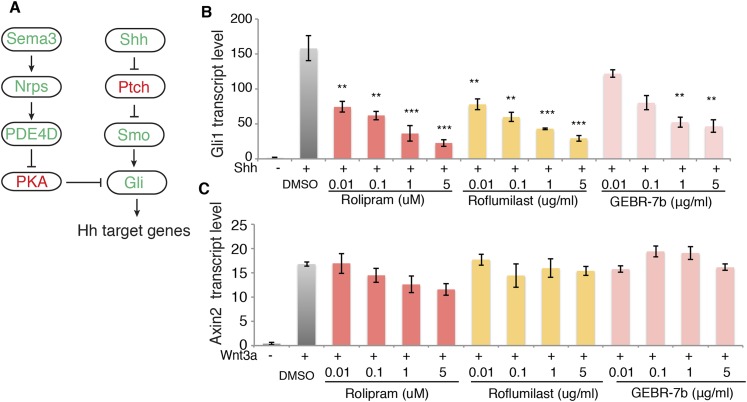
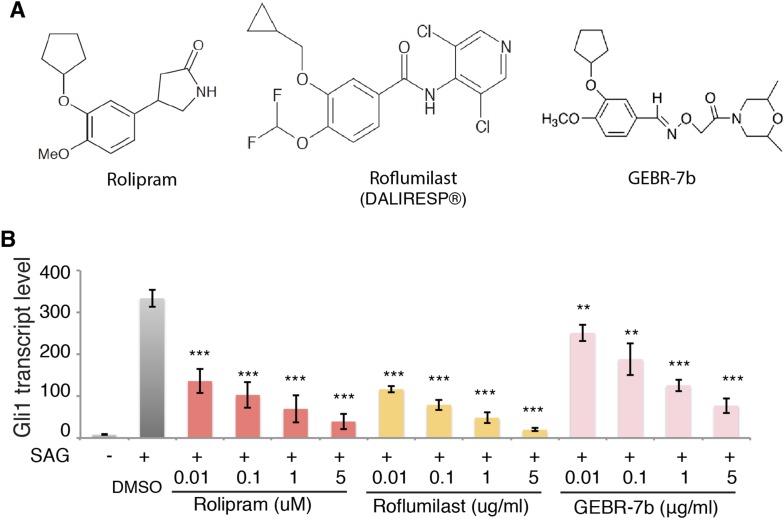
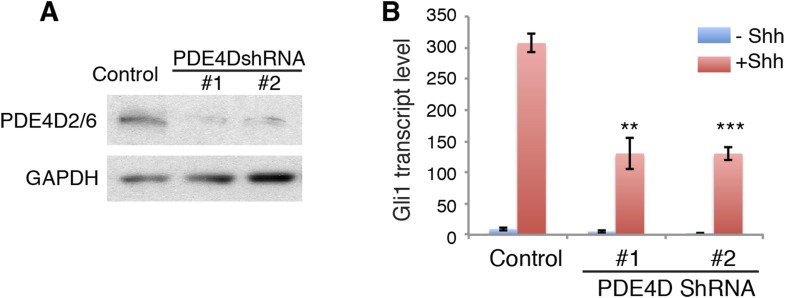
Figure 4. PDE4D inhibitors suppress Hh signal transduction.
(A) Schematic diagram showing that Sema3-Nrp signaling regulates Hh pathway through controlling the activity of PDE4D and PKA. (B) The Hh signaling activity in NIH3T3 cells incubated with PDE4D inhibitors together with Shh conditioned medium for 4 hr Gli1 transcript levels were measured by qPCR to evaluate the Hh signal transduction. (C) The Wnt signaling activity in NIH3T3 cells incubated with PDE4D inhibitors together with Wnt3a for 4 hr Axin2 transcript levels were measured by qPCR to evaluate the Wnt signal transduction. All data are mean ± SEM. Statistics: Student t-Test, in comparison to the condition where cells were treated with DMSO. **p < 0.01, ***p < 0.001.