Abstract
Heat shock transcription factors (Hsfs) are known to play dominant roles in plant responses to heat, as well as other abiotic or biotic stress stimuli. While the strawberry is an economically important fruit plant, little is known about the Hsf family in the strawberry. To explore the functions of strawberry Hsfs in abiotic and biotic stress responses, this study identified 17 Hsf genes (FvHsfs) in a wild diploid woodland strawberry (Fragaria vesca, 2n = 2x = 14) and isolated 14 of these genes. Phylogenetic analysis divided the strawberry FvHsfs genes into three main groups. The evolutionary and structural analyses revealed that the FvHsf family is conserved. The promoter sequences of the FvHsf genes contain upstream regulatory elements corresponding to different stress stimuli. In addition, 14 FvHsf-GFP fusion proteins showed differential subcellular localization in Arabidopsis mesophyll protoplasts. Furthermore, we examined the expression of the 17 FvHsf genes in wild diploid woodland strawberries under various conditions, including abiotic stresses (heat, cold, drought, and salt), biotic stress (powdery mildew infection), and hormone treatments (abscisic acid, ethephon, methyl jasmonate, and salicylic acid). Fifteen of the seventeen FvHsf genes exhibited distinct changes on the transcriptional level during heat treatment. Of these 15 FvHsfs, 8 FvHsfs also exhibited distinct responses to other stimuli on the transcriptional level, indicating versatile roles in the response to abiotic and biotic stresses. Taken together, the present work may provide the basis for further studies to dissect FvHsf function in response to stress stimuli.
Keywords: strawberry (Fragaria vesca), heat shock transcription factor, expression analysis, subcellular localization, heat stress, abiotic stress, phytohormones
Introduction
Global warming has brought about many severe environmental problems with adverse impacts on almost all aspects of plant development, growth, reproduction, or yield (Hedhly et al., 2009; Mittler et al., 2012). Although plants are sessile organisms that cannot escape from stress conditions, a cascade of activities regulated by transcription factors are triggered in response to such conditions (Schwechheimer and Bevan, 1998). Particularly, plant heat shock transcription factors (Hsfs) are known to play a central role in protecting plants from heat or other stress conditions (Scharf et al., 2012).
Hsfs are structurally conserved throughout the eukaryotic kingdom. Earlier studies focusing on the structural and biochemical characteristics of Hsfs (Harrison et al., 1994; Littlefield and Nelson, 1999) revealed that a typical Hsf consists of five conserved motifs, including a DNA-binding domain (DBD), an oligomerization domain (OD), a nuclear localization signal (NLS), a nuclear export signal (NES), and an activator peptide motif (AHA) (Never et al., 1996; Scharf et al., 2012).
A number of studies have demonstrated that Hsfs play vital roles in plant responses to abiotic stresses such as heat, cold, salt, drought, and oxidative conditions, among others (Schramm et al., 2008; Yoshida et al., 2010; Hwang et al., 2014). For example, a versatile regulatory regime involving HsfA1, HsfA2, HsfB1a, Hsp90, and Hsp70 in the control of heat stress response was proposed (Hahn et al., 2011). HsfA2 serves as the most strongly induced Hsf, accumulating to high levels after exposure of tomato (Chan-Schaminet et al., 2009), Arabidopsis (Schramm et al., 2006), or rice (Mittal et al., 2009) to long-term heat stress. Substantial evidence has also demonstrated that HsfA2 is associated with the expression of multiple Hsps or general stress-related non-chaperone encoding genes (Schramm et al., 2006; Chan-Schaminet et al., 2009; Nishizawa-Yokoi et al., 2009). HsfA3 is located downstream of the DREB2A stress-regulatory system in the transcriptional cascade (Schramm et al., 2008) and could be involved in physiological responses to drought and salt stress (Li et al., 2013). HsfA4 was reported to be involved in oxidative stress (Davletova et al., 2005) and cadmium tolerance in rice and wheat (Shim et al., 2009). HsfA6a and HsfA6b expression is highly increased under high salinity or dehydration conditions (Yoshida et al., 2010; Hwang et al., 2014). In addition, HsfB1a and HsfB2a/b were reported to be associated with pathogen resistance in Arabidopsis (Kumar et al., 2009; Ikeda et al., 2011; Pick et al., 2012). In addition to stress resistance, Hsfs have also been linked to important roles in plant growth and development (Kotak et al., 2007; Almoguera et al., 2009). Recently, genome-wide expression profiles analyses in rice (Mittal et al., 2009), wheat (Xue et al., 2014), soybean (Chung et al., 2013), and apple (Giorno et al., 2012) also indicated that several Hsf genes were transcribed at high levels during heat, cold, salt, and drought stresses. However, little is known about Hsf genes in the strawberry.
The strawberry is one of the most important fruit crops in the world. However, more extreme temperatures and droughts, serious fungal infections and secondary salinization have strongly limited the growth, development, reproduction, and yield of strawberry plants in recent years (Maughan et al., 2015; Nezhadahmadi et al., 2015). The woodland strawberry F. vesca has a smaller genome (~240 Mb) that is highly congenic with its octoploid cultivated species (Fragaria × ananassa Duch.) (Shulaev et al., 2011). These features underlie its potential utility as a versatile experimental system for studying the counterparts of many important genes in Rosaceae fruit crops (Shulaev et al., 2011; Liu et al., 2014).
To explore the functions of Hsfs in abiotic and biotic stress responses in the strawberry, the current study identified and isolated FvHsf genes in a diploid woodland strawberry accession Heilongjiang-3 (Liu et al., 2014), in addition to analyzing the evolutionary relationships, gene structure, protein domains, and expression profiles of these genes with respect to heat, drought, salt, cold, and powdery mildew infection stresses and abscisic acid, ethephon, methyl jasmonate, and salicylic acid treatments. Notably, the subcellular localization of most (14/17) of the FvHsf proteins was demonstrated; these data have not been fully elucidated in any other plant species. Taken together, the present work may provide the basis for further studies to dissect FvHsf function in response to abiotic or biotic stresses.
Materials and methods
Identification and classification of Hsf genes in strawberry
A publicly available database (http://planttfdb.cbi.pku.edu.cn/) predicted that 15 Hsf genes exist in the diploid strawberry genome (accession Hawaii-4) and predicted the corresponding nucleotide and amino acid sequences. All of the nucleotide and amino acid sequences of the predicted FvHsf genes were used as queries to perform BLAST searches in the public NCBI database. The strawberry genes with the highest identity (>90%) and lowest E-value (0) were selected as candidates. If two or more protein sequences at the same gene locus overlapped, only the longest sequence was used. All of the putative candidates were manually verified with the InterProScan program (http://www.ebi.ac.uk/Tools/pfa/iprscan/) to confirm their completeness and the presence of a DNA-binding domain (PF00447) (http://pfam.sanger.ac.uk/).
Phylogenetic analysis was used to classify the FvHsf genes into three classes and further subclasses (Scharf et al., 2012). The full-length amino acid sequences of Hsf proteins from strawberry (FvHsf), A. thaliana (AtHsf), rice (Oryza sativa L., OsHsf) (Guo et al., 2008), apple (Malus domestica Borkh, MdHsf) (Giorno et al., 2012), and grape (Vitis vinifera, VvHsf) were used to generate a phylogenetic tree through ClustalW alignment and the unrooted Neighbor-joining method using MEGA 5.0 (Tamura et al., 2011). Neighbor-joining analysis with pairwise deletion was performed using the Jones-Taylor-Thornton (JTT) model. Bootstrap analysis was performed with 1000 replicates to assess the level of statistical support for each tree node (Xue et al., 2014).
Gene structure, protein domains, and synteny analysis of strawberry Hsf genes
The exon-intron structures of FvHsf genes were visualized using the online program GSDS 2.0 (http://gsds.cbi.pku.edu.cn/), which aligns the respective coding sequences with corresponding full-length sequences. The protein domains of the FvHsf genes were identified using MEME online tools (http://meme.nbcr.net/meme/) with the parameters set as follows: the minimum length of the conserved motif was 6, the maximum length of the conserved motif was 100, and the largest number of discovered and conserved motifs was 25. The other parameters retained their default settings. The results were presented by DOG 1.0 (Ren et al., 2009). The syntenic blocks used to construct a synteny analysis map between the strawberry and Arabidopsis Hsf genes were obtained from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/) and Plant Genome Duplication Database (Lee et al., 2013), and the diagrams were generated by the Circos program, version 0.63 (http://circos.ca/).
Isolation and subcellular localization of strawberry Hsf genes
To provide more insight into the function of FvHsfs, we designed gene-specific primers to isolate the putative Hsf genes from a diploid woodland strawberry accession Heilongjiang-3. The predicted full-length coding sequences of FvHsf genes were amplified from Heilongjiang-3 cDNA using high-fidelity Taq HS-mediated PCR. Then, we used the isolated FvHsfs to examine the subcellular localization of these proteins. The amplified PCR products were digested with XbaI/XhoI and KpnI and fused in-frame with GFP in the XbaI/XhoI and KpnI site of the pBI221 vector containing the CaMV 35S promoter (Clontech, Beijing, China), resulting in the pFvHsf-GFP plasmids. The primers used to clone genes and construct vectors are available in Supplementary Table S1.
For the transient expression of FvHsfs-GFP in Arabidopsis mesophyll protoplasts, the corresponding pFvHsfs-GFP plasmid was transformed into Arabidopsis mesophyll protoplasts using the PEG-calcium transfection method described in Yoo et al. (2007). After transformation, the Arabidopsis mesophyll protoplasts were kept in darkness at room temperature for 16–18 h before examination by fluorescence microscopy. Images were acquired using an Olympus BX-51 inverted fluorescence microscope (Olympus, Japan). The image data were processed using Adobe Photoshop (Mountain View, CA, USA). All of the transient expression assays were repeated at least three times.
In silico promoter analysis
The cis-elements presented in the 1 kb region upstream of the translation start site of the strawberry Hsf genes were obtained from the PlantCARE database (Rombauts et al., 1999), and the corresponding nucleotide sequences were retrieved from NCBI with their gene IDs (http://www.ncbi.nlm.nih.gov/).
Plant materials and treatments
The wild diploid strawberry F. vesca accession Heilongjiang-3 was grown in the strawberry germplasm resource greenhouse of Northwest A&F University. The potted strawberry plants were grown at 22°C with 70% relative humidity and no supplemental light. Six-month-old strawberry seedlings with the tenth leaf fully expanded were selected for treatments. The roots, stems, leaves, flowers, receptacles, and fruits of greenhouse-grown Heilongjiang-3 were used to analyze the organ-specific expression of the FvHsf genes. A. thaliana ecotype Col-0 was grown at 22°C with 75% relative humidity under short-day (8 h light at 125 μmol ·m−2·s −1, 16 h dark) conditions for 4–5 weeks before transformation.
Heat stress treatment was performed by transferring potted strawberry plants to a 42°C chamber for 48 h. Leaves were collected from the plants at 0, 0.5, 2, 4, 8, 12, 24, and 48 h post-treatment (hpt). Cold stress treatment was performed by transferring the plants to a 4°C chamber for 48 h. Leaves were harvested from the plants at 0, 0.5, 2, 4, 8, 12, 24, and 48 hpt. For both the heat and cold treatments, another set of potted Heilongjiang-3 seedlings were kept in the control temperature range of 22–27°C. Salt stress was simulated by irrigating potted strawberry plants with 300 mM NaCl once. Another set of control Heilongjiang-3 seedlings was similarly treated with distilled water. The leaves were collected from the plants at 0, 0.5, 2, 4, 8, 12, 24, and 48 hpt. Drought stress was simulated by withholding water and sampling at 0, 24, 48, 72, 96, 120, and 144 hpt. The plants were rewatered after 144 h of drought stress and sampled again 24 h later. The plants were inoculated with powdery mildew (Podosphaera aphanis) by touching the adaxial epidermis of Heilongjiang-3 with sporulating colonies located on the leaf surface of the strawberry cv. Red cheeks. Another set of control Heilongjiang-3 seedlings was similarly touched with uninfected, healthy leaves. The treated plants were then incubated in a controlled environment. Inoculated leaves were collected at 0, 24, 48, 72, 96, 120, 144, and 168 h post-inoculation (hpi). The inoculations were repeated three times. For hormone treatments, the strawberry leaves were sprayed with a solution containing 0.1 mM abscisic acid (ABA), 1 mM salicylic acid (SA), 0.1 mM methyl jasmonate (MeJA), or 0.5 g/L ethephon (Eth), while another set of control Heilongjiang-3 seedlings were similarly sprayed with distilled water. The treated leaves were then collected at 0, 0.5, 2, 4, 8, 12, 24, and 48 hpt for RNA isolation. Six leaves from six separate plants were collected at each time point of each treatment and combined to form one sample, and all of the experiments were performed independently in triplicate.
Semi-quantitative PCR and real-time quantitative PCR analysis
Total RNA was isolated from treated leaves or tissue samples using an EZNA Plant RNA Kit (R6827-01, Omega Bio-tek, USA). cDNA synthesis was performed using PrimeScript RTase (TaKaRa Biotechnology, Dalian, China). The strawberry 18S gene was used as a control to normalize the amount of cDNA used from each sample (Raab et al., 2006). VECTOR NTI was used to design gene-specific primers for each FvHsf gene (Supplementary Table S1). The following semi-quantitative reverse-transcription PCR program was used: 95°C for 3 min, 30–32 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s, and a final step of 72°C for 5 min. The PCR products were separated on a 1.0% (w/v) agarose gel, stained with ethidium bromide, and imaged under UV light for further gene expression analysis. Each reaction was performed in triplicate, with the three independent analyses for each treatment showing the same trends for each gene. The expression profiles from the semi-quantitative RT-PCR were collated, analyzed, and visualized using the GeneSnap and MeV 4.8.1 programs.
Real-time quantitative PCR was carried out using SYBR green (TaKaRa Biotechnology) on an IQ5 real time-PCR machine (Bio-Rad, Hercules, CA, USA) with a final volume of 21 μl per reaction. Each reaction mixture contained 10.5 μl SYBR Premix Ex Taq II (TaKaRa Biotechnology), 1.0 μl cDNA template, 1.0 μl of each primer (1.0 μM), and 7 μl sterile distilled H2O. Each reaction was performed in triplicate. The cycling parameters were 95°C for 30 s, 40 cycles at 95°C for 30 s and 58°C for 30 s. After amplification, the samples were kept at 50°C for 1 min and the temperature was gradually raised by 0.5°C every 10 s to perform the melt-curve analysis. Relative gene expression was determined using an 18S-26S interspacer gene as an endogenous control gene (Raab et al., 2006). Each relative expression level was analyzed with IQ5 software using the Normalized Expression method. The primers used for real-time RT-qPCR are listed in Supplementary Table S1.
Statistical analysis
The data are presented as the mean ± standard deviation of the mean (SD) and were tested for statistical significance with a paired Student's t-test (http://www.physics.csbsju.edu/stats/). p < 0.05 was selected as the point of minimal statistical significance in all of the analyses.
Results
Identification and isolation of Hsf genes in the diploid strawberry
A total of 17 strawberry Hsf genes were originally obtained through the BLAST search in NCBI using the 15 strawberry Hsf amino acid sequences that were predicted in the sequenced genome of the accession Hawaii-4 (http://planttfdb.cbi.pku.edu.cn/) (Shulaev et al., 2011). All 17 of the proteins contained apparently complete Hsf-type DNA-binding domains (PF00447) and oligomerization domains (Never et al., 1996; Scharf et al., 2012). As a result, 17 FvHsf members were identified in the diploid strawberry (Table 1). More detailed information about each FvHsf gene, including the Hsf gene IDs, gene location, length of the coding sequences, and the characteristics of the FvHsf proteins can be found in Table 1. Using the predicted FvHsf coding sequences, we initially isolated 14 homologous genes from a diploid woodland strawberry accession Heilongjiang-3 (F. vesca). It is notable that the ORF sequences of the isolated FvHsfs from accession Heilongjiang-3 share high identities (≥96%, E-value = 0) with the corresponding FvHsfs in strawberry accession Hawaii-4 (Shulaev et al., 2011) (Table 1).
Table 1.
Characteristics of strawberry Hsf genes.
| Name | Gene IDa | Gene location | Length (bp) | Number of aa | MWb (kDa) | Isoelectric pointb | Identity/ E-valuec | Accession numbersd |
|---|---|---|---|---|---|---|---|---|
| FvHsfA1b | 101302336 | LG3: 11614311–11618187 | 3876 | 494 | 55.01 | 5.02 | 99%, 0.0 | KT283219 |
| FvHsfA1d | 101302657 | LG5:21020179–21025194 | 5015 | 508 | 55.66 | 4.80 | 99%, 0.0 | KT283220 |
| FvHsfA2a | 101312795 | LG2:20570433–20572945 | 2512 | 372 | 41.97 | 4.86 | 99%, 0.0 | KT283221 |
| FvHsfA3a | 101295258 | LG6:13129046–13132026 | 2980 | 456 | 50.35 | 4.77 | 99%, 0.0 | KT283222 |
| FvHsfA4a | 101311435 | LGun:147013–149497 | 2484 | 497 | 55.72 | 5.13 | 99%, 0.0 | KT283223 |
| FvHsfA4b | 101290970 | LG6:16216378–16219992 | 3614 | 439 | 49.77 | 4.99 | – | – |
| FvHsfA5a | 101302160 | LG4:14062542–14066187 | 3645 | 578 | 64.93 | 7.40 | 99%, 0.0 | KT283224 |
| FvHsfA6a | 101307359 | LG2:16115738–16118875 | 3137 | 362 | 41.18 | 5.79 | 99%, 0.0 | KT283225 |
| FvHsfA7a | 101297339 | LG4:5513754–5515879 | 2125 | 345 | 40.13 | 5.42 | – | – |
| FvHsfA8a | 101290755 | LG4:19035204–19038162 | 2958 | 474 | 54.45 | 4.99 | – | – |
| FvHsfA9a | 101302973 | LG1:6313891–6316458 | 2567 | 447 | 50.90 | 5.23 | 99%, 0.0 | KT283226 |
| FvHsfB1a | 101294882 | LG1:10956423–10958906 | 2483 | 290 | 32.01 | 5.96 | 99%, 0.0 | KT283227 |
| FvHsfB2a | 101299242 | LG7:22683186–22684501 | 1315 | 289 | 31.97 | 8.82 | 98%, 0.0 | KT283228 |
| FvHsfB2b | 101301205 | LG5:1089695–1092781 | 3086 | 355 | 38.41 | 4.79 | 96%, 0.0 | KT283229 |
| FvHsfB3a | 101307517 | LG6:33122976–33124719 | 1743 | 236 | 27.10 | 5.91 | 99%, 0.0 | KT283230 |
| FvHsfB4a | 101301353 | LG2:21014849–21016923 | 2074 | 386 | 43.21 | 7.73 | 99%, 0.0 | KT283231 |
| FvHsfC1a | 101304412 | LG1:3848442–3850066 | 1624 | 362 | 40.49 | 5.77 | 99%, 0.0 | KT283232 |
IDs are available in the strawberry Genome Database (http://www.rosaceae.org).
Detailed FvHsf protein characteristics were predicted by ExPASy online service (http://web.expasy.org/cgi-bin/compute_pi/pi_tool).
The identities of the isolated FvHsf genes were derived from a NCBI Blast search using the FvHsf Heilongjiang-3 nucleotide sequences.
Sequence data of isolated FvHsfs can be found in GenBank databases using these accession numbers.
Phylogenetic analysis and classification of FvHsf proteins
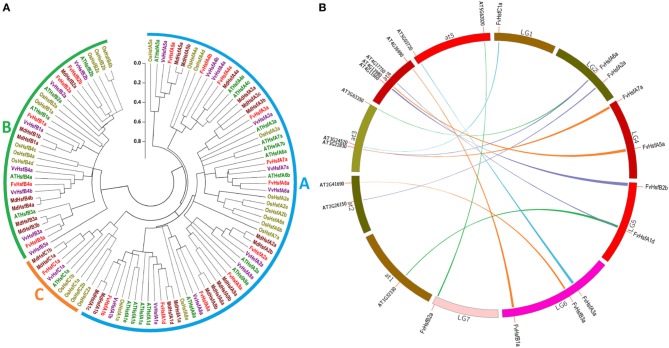
Compared with fungi and animals, plants possess large numbers of Hsf genes (Von Koskull-Doring et al., 2007), and the plant Hsf genes were assigned into A, B, and C classes based on the diversity of the oligomerization domain (Never et al., 1996). To examine the classification of FvHsf members and to gain some insight into the potential function of FvHsf proteins from well-studied Hsfs in other plant species, we used full-length amino acid sequences to perform a phylogenetic analysis of Hsf proteins from strawberry, rice, apple, grapevine, and Arabidopsis (Figure 1A). The phylogenetic analysis indicated that the 106 Hsf proteins from these plants were clearly grouped into three different clades corresponding to the Hsf classes A, B, and C (Figure 1A, Table 1, and Supplementary Table S2). Within each Hsf protein class, particular clusters of orthologous and paralogous genes have been identified, showing ancestral speciation or duplication events. For example, the monocot rice has an additional C2 clade but has lost the A9 and B3 clades (Figure 1A). Although all of the clades resolved in dicots (apple, grapevine, and Arabidopsis) are represented in the strawberry Hsf proteins, fewer paralogous pairs may limit the size of the FvHsf family (Figure 1A).
Figure 1.
Comparative analysis of strawberry Hsf genes with the corresponding genes in rice, apple, grapevine, and Arabidopsis. (A) The linearized neighbor-joining phylogenetic tree of Hsf proteins from strawberry, rice, apple, grapevine, and Arabidopsis. The full-length amino acid sequences of the Hsf proteins were used to construct the phylogenetic tree using the MEGA 5.0 program. Unrooted neighbor-joining analysis was performed with pairwise deletion and the Jones-Taylor-Thornton (JTT) model. The filled circle lines were used to cluster the genes into the A, B, and C classes. (B) Synteny analysis of strawberry and Arabidopsis Hsf genes. The strawberry and Arabidopsis chromosomes are depicted as a circle. The approximate location of each AtHsf and FvHsf gene is marked with a short red line on the circle. The colored curves denote the syntenic regions of the strawberry and Arabidopsis Hsf genes.
Syntenic relations, exon-intron organization, and protein domains of the FvHsf genes
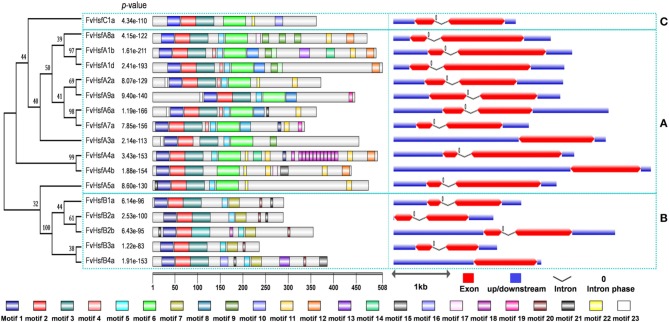
Most of the Arabidopsis Hsf genes have been systematically investigated in recent years, and a synteny analysis of strawberry Hsfs and Arabidopsis Hsfs was performed in the present work to ascertain whether this information might provide more functional insight. A total of 11 pairs of syntenic Hsf genes were found between strawberry and Arabidopsis; this number included 11 FvHsf genes and 12 AtHsf genes (Supplementary Table S3; Figure 1B). Interestingly, two FvHsf genes (FvHsfA1d, FvHsfA6a) and one AtHsf gene (AtHsfA6b) were found to be associated with two syntenic blocks (Supplementary Table S3). Insights into the structure of the FvHsf genes were obtained through an analysis of the exon/intron boundaries, which are known to play key roles in gene family evolution (Schwartz et al., 2009). As shown in Figure 2, the FvHsf genes exhibit a highly conserved exon-intron organization: of the 17 FvHsf genes, 14 possess two exons and three have one exon. It is interesting that the introns of the FvHsf genes divide the highly conserved DNA-binding domains coding regions into two parts, and all of the introns are phase zero introns. Plant Hsf proteins show a highly modular assembly of protein domains (Never et al., 1996; Scharf et al., 2012). We used the online MEME tool to predict the conserved FvHsf protein domains, identifying 23 conserved motifs (Figure 2; Supplementary Table S2). Unsurprisingly, all 17 of the FvHsfs showed the presence of a DNA-binding domain (DBD) in the N-terminal of the protein (Figure 2; Supplementary Table S2). The oligomerization domains (OD or HR-A/B region), which differ in sequence among the class A, B, and C members, are adjacent to the DBD (Figure 2; Supplementary Table S2). In addition, nuclear localization signal (NLS) motifs were found in 15 FvHsfs, nuclear export signal (NES) motifs were found in five class A FvHsfs, and transcription activator (AHA) motifs were found in eight class A FvHsfs (Figure 2; Supplementary Table S2). Overall, each FvHsf protein contained the necessary DBD and OD, but some FvHsfs have specific motifs such as NLS, NES, and/or AHA, which might lay the foundation for the functional divergences between different FvHsfs (Scharf et al., 1998; Döring et al., 2000; Heerklotz et al., 2001).
Figure 2.
Structural analysis of strawberry Hsf genes. The protein domains of the strawberry Hsf genes are shown on the left and are denoted by rectangles with different colors. The exon-intron organization is shown on the right, with exons and introns represented by red wedges and black lines, respectively, and untranslated regions (UTRs) indicated by blue boxes. The number 0 represents the intron phase. The exon size can be estimated using the scale at the bottom of the diagram, while all of the introns were set to same length. The blue dashed rectangles are used to cluster the genes into the A, B, and C classes.
Regulatory elements in the FvHsf promoters
An in silico survey of the putative cis-elements in the 1 kb region upstream of the translation initiation codon of various FvHsf genes showed the presence of some abiotic stress response elements such as HSEs (heat shock elements), which were found in FvHsfA2a, FvHsfA4a, FvHsfA6a, FvHsfB2a, and FvHsfC1a (Figure 3). In addition, LTR, a cis-acting element involved in low-temperature responsiveness (Jiang et al., 1996) was observed in FvHsfA1b, FvHsfA4a, and FvHsfA8a; anoxia responsive elements (ARE) were observed in 13 FvHsf genes; MBS (MYB binding site), a cis-acting element involved in drought-inducibility (Deng et al., 1996), was found in 10 FvHsf genes; Box-W1, a fungal elicitor responsive element, was identified in FvHsfA5a, FvHsfA9a, and FvHsfB2a; and TC-rich repeats, a cis-acting element involved in defense and stress responsiveness, was seen in 11 FvHsf genes. Lastly, ABA responsive elements (ABREs) were found in nine FvHsf genes, MeJA responsive elements (CGTCA-motif/TGACG-motif) were observed in eight FvHsf genes, and SA responsive elements (TCA-element) were seen in nine FvHsf genes (Figure 3). Notably, plenty of hormone-responsive elements were seen in the FvHsf promoter sequences, indicating that phytohormones could play central roles in regulation of heat stress responses (Clarke et al., 2009).
Figure 3.
Cis-elements are present in the 1 kb region upstream of the strawberry Hsf translation start site. The analysis was performed using the PlantCARE database. The different-colored boxes mark the relative positions of the different elements.
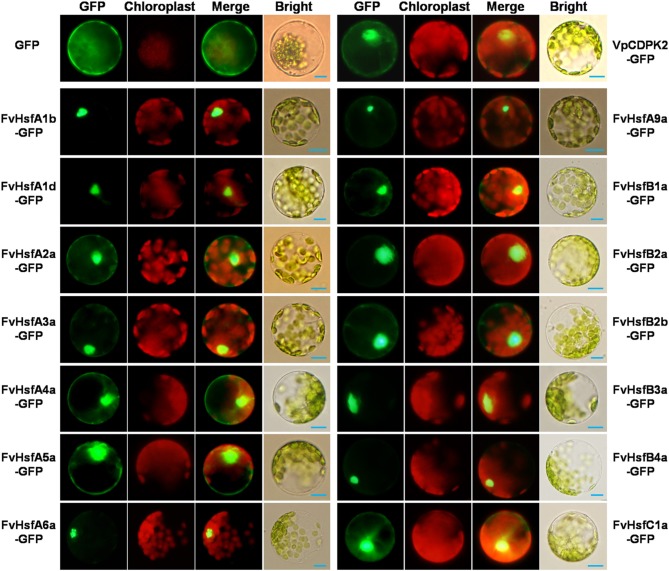
Subcellular localization of FvHsfs
The cellular localization of Hsfs is essential to their functions (Scharf et al., 1998). To investigate the subcellular localization of FvHsfs, the coding-sequences of 14 FvHsf genes were fused in-frame with GFP under control of the CaMV 35S promoter. The fusion constructs and the positive control were transiently expressed in Arabidopsis mesophyll protoplasts. As shown in Figure 4, the GFP expressed from the control construct was dispersed throughout the whole cell, whereas the most of the FvHsf fusion proteins were clearly located in the nucleus. However, some of the FvHsf fusion proteins, including FvHsfA2a-GFP, FvHsfA3a-GFP, FvHsfA4a-GFP, FvHsfA5a-GFP, FvHsfB2b-GFP, and FvHsfC1a-GFP, were also detected in the cytosol. To confirm the nuclear and cytosolic localization of the FvHsfs, we also examined the subcellular localization of VpCDPK2, a grape calcium-dependent protein kinase that was reported to localize to both the nucleus and cytosol of Arabidopsis mesophyll protoplasts (Zhang et al., 2015). The FvHsfs and VpCDPK2 exhibited similar subcellular distributions (Figure 4). These results might have broader implications for understanding the subcellular localization of plant Hsfs (Lyck et al., 1997; Kotak et al., 2004).
Figure 4.
The subcellular localization of 14 FvHsfs. The selected Hsf genes were cloned from a diploid woodland strawberry (F. vesca) and used to construct CaMV35S::Hsfs-GFP vectors in which GFP was fused at the C terminus. The 14 FvHsf-GFP fusion proteins (FvHsfA1b-GFP, FvHsfA1d-GFP, FvHsfA2a-GFP, FvHsfA3a-GFP, FvHsfA4a-GFP, FvHsfA5a-GFP, FvHsfA6a-GFP, FvHsfA9a-GFP, FvHsfB1a-GFP, FvHsfB2a-GFP, FvHsfB2b-GFP, FvHsfB3a-GFP, FvHsfB4a-GFP, and FvHsfC1a-GFP), the VpCDPK2-GFP marker protein, and GFP control were transiently expressed in A. thaliana mesophyll protoplasts and observed by fluorescence microscopy. The merged pictures include the green fluorescence channel (first panels) and the chloroplast autofluorescence channel (second panels). The corresponding bright field images are shown on the right. Bars = 10 μm.
Expression profiles of FvHsfs in different organs of the strawberry plant
To determine the biological roles of Hsf genes in strawberry Heilongjiang-3, the distribution of 17 FvHsfs transcripts was surveyed in six major organs (roots, stems, leaves, flowers, receptacles, and fruits) under non-stress conditions. As shown in Figure 5B, the FvHsfA1d, FvHsfA2a, and FvHsfB3a transcripts showed consistent distribution throughout all of the tested organs, and most of the FvHsf genes exhibited high mRNA levels in the fruits of Heilongjiang-3, with the exception of FvHsfB2a and FvHsfC1a. The FvHsfA1b, FvHsfA4b, FvHsfA5a, FvHsfB1a, FvHsfB2b, and FvHsfC1a genes showed high transcription levels in the flowers and receptacles of Heilongjiang-3; FvHsfA1b, FvHsfA3a, FvHsfA4a, FvHsfA6a, and FvHsfC1a also exhibited higher transcript abundance in old leaves than in roots, stems, and young leaves. In addition, the FvHsfA1b, FvHsfA4b, FvHsfA5a, FvHsfA8a, FvHsfA9a, and FvHsfB1a transcripts could not be detected in the roots of Heilongjiang-3, while FvHsfB2a could not be detected in any Heilongjiang-3 tissues. The organ-specific FvHsf expression patterns suggest that FvHsfs could play important biological roles in the growth and development of strawberries (Kotak et al., 2007).
Figure 5.
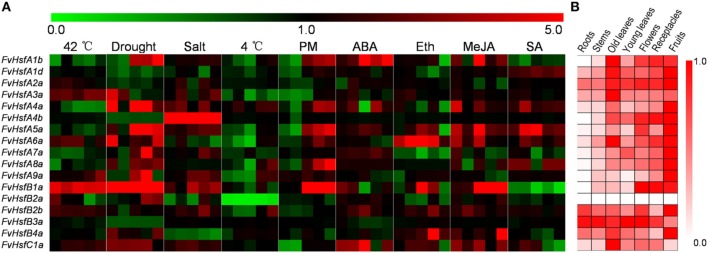
The expression pattern of 17 FvHsf genes in different organs/tissues and under different treatments. The expression profiles were generated by semi-quantitative PCR and were visualized as heat maps. The color scale represents log2 expression values, with red denoting increased transcript abundance and green denoting decreased transcript abundance. Only partial data are shown for the stress and hormone treatments (the expression levels of 17 FvHsfs at 0.5 hpt, 2 hpt, 4 hpt, 24 hpt, and 48 hpt of heat, cold, salt, and hormone treatments; the expression levels of 17 FvHsfs at 24 hpt/i, 48 hpt/i, 72 hpt/i, 144 hpt/i, and 168 hpt/i of drought treatment and powdery mildew infection). Detailed expression profiles of all 17 FvHsfs in response to all treatments are provided in Supplementary Figures S1, S2. (A) The expression profiles of the 17 FvHsfs in response to different treatments. (B) The organ/tissue distribution of 17 FvHsf genes under homeostatic conditions, with the red marks indicating the relative level of each FvHsf transcript in different organs/tissues.
FvHsfs act as positive responders to heat stress in strawberry
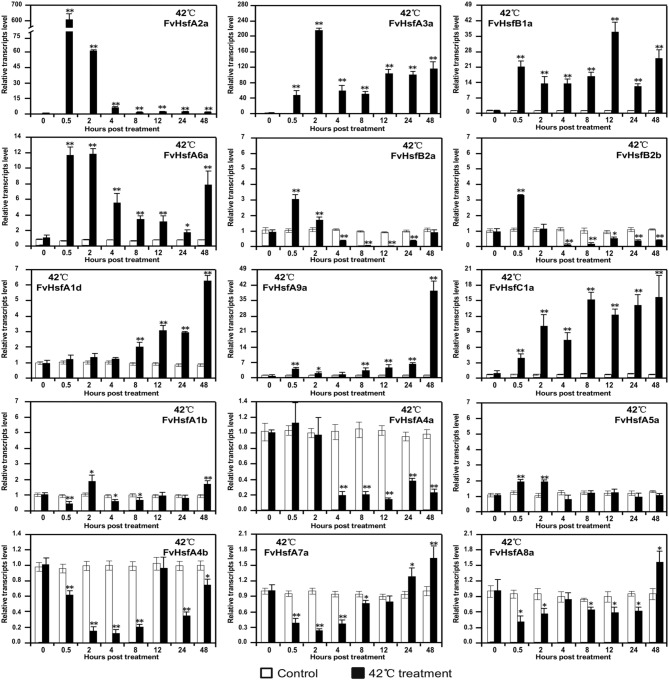
To understand how Hsf genes respond to heat stress in strawberry, we exposed Heilongjiang-3 to 42°C conditions and performed semi-quantitative RT-PCR to determine expression profiles for the FvHsf gene family (Figure 5A and Supplementary Figure S1). Based on the results of the semi-quantitative RT-PCR, we selected 15 characteristic FvHsfs using real-time quantitative RT-PCR to further test their transcript abundance during 42°C treatment (Figure 6).
Figure 6.
The expression of FvHsf genes in the diploid woodland strawberry (F. vesca) in response to 42°C treatment. Real-time quantitative PCR was performed to determine the expression of various FvHsfs in 42°C temperatures relative to control temperatures. The analysis results were normalized using Fv18s. The experiments were repeated three times and gave consistent results. The mean values and SDs were obtained from three biological and three technical replicates. The asterisks indicate that the corresponding gene was significantly up or down-regulated in response to treatment, as determined by the Student's t-test (*P < 0.05, **P < 0.01).
As shown in Figure 6, the 15 FvHsf genes showed distinct expression patterns during 42°C treatment. Of the 15 FvHsfs, FvHsfA2a was the most strongly heat stress induced Hsf gene in strawberry, reaching its highest transcription level (~600-fold) at 0.5 hpt. FvHsfA3a, FvHsfA6a, and FvHsfB1a were also strongly induced at 0.5 hpt, maintaining elevated mRNA levels (~10- to 200-fold) throughout the entire treatment period. FvHsfA1d, FvHsfA9a, and FvHsfC1a were highly induced at the late stage (24–48 hpt) of 42°C treatment, with these transcripts up-regulated 6- to 40-fold during this time period. FvHsfA5a, FvHsfB2a, and FvHsfB2b were up-regulated in the earlier stage of treatment (0.5–2 hpt), but returned to baseline or were down-regulated after 2 hpt. FvHsfA4a, FvHsfA4b, FvHsfA7a, and FvHsfA8a were primarily down-regulated in response to treatment, but FvHsfA7a and FvHsfA8a returned to baseline at the late stage of treatment. FvHsfA1b was only slightly up-regulated at 2 and 48 hpt.
FvHsfs are associated with other abiotic and biotic stresses in strawberry
Although Hsfs are primarily involved in heat acclimatization, these genes have also been reported to play roles in plant adaptations to other environmental stresses, including cold (Mittal et al., 2009), salt (Hwang et al., 2014), drought (Prieto-Dapena et al., 2008; Hwang et al., 2014) and pathogen infection (Pick et al., 2012). In this study, the semi-quantitative RT-PCR indicated that most of the FvHsfs (FvHsfA1b, A2a, A3a, A4a, A5a, A6a, A8a, A9a, B1a, B2a, B2b, B4a, and C1a) showed detectable changes at the transcript level when strawberry plants were exposed to specific stress conditions (Figure 5A; Supplementary Figures S1, S2). These data suggest that these FvHsfs are involved in the response to almost all stress treatments, but examination with real-time quantitative RT-PCR revealed that the expression of each FvHsf is highly specific (Figures 7, 8).
Figure 7.
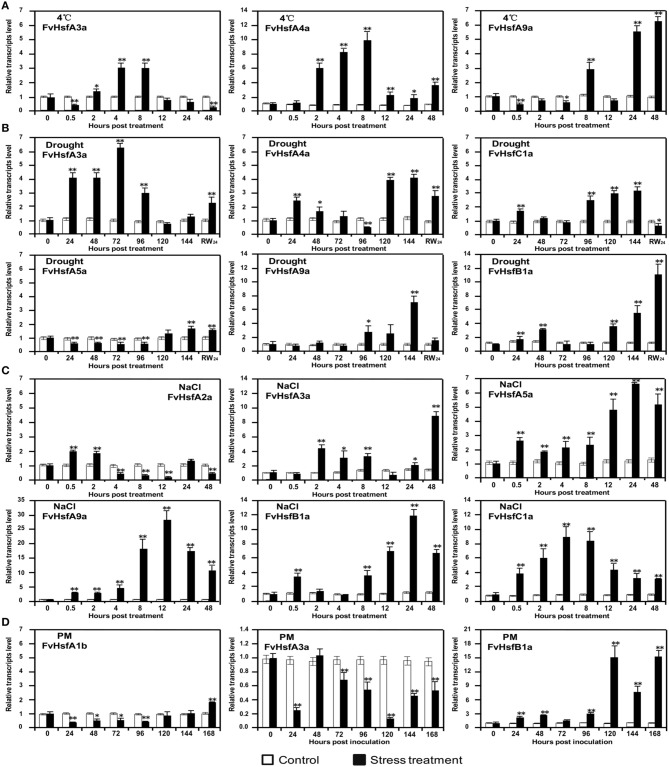
The expression of Hsf genes in the diploid woodland strawberry (F. vesca) in response to 4°C, drought, salt, and Podosphaera aphanis (PM) treatments. The expression levels of several representative FvHsf genes showed unusual patterns in response to 4°C (A), drought (B), salt (C), and PM (D) treatments.
Figure 8.
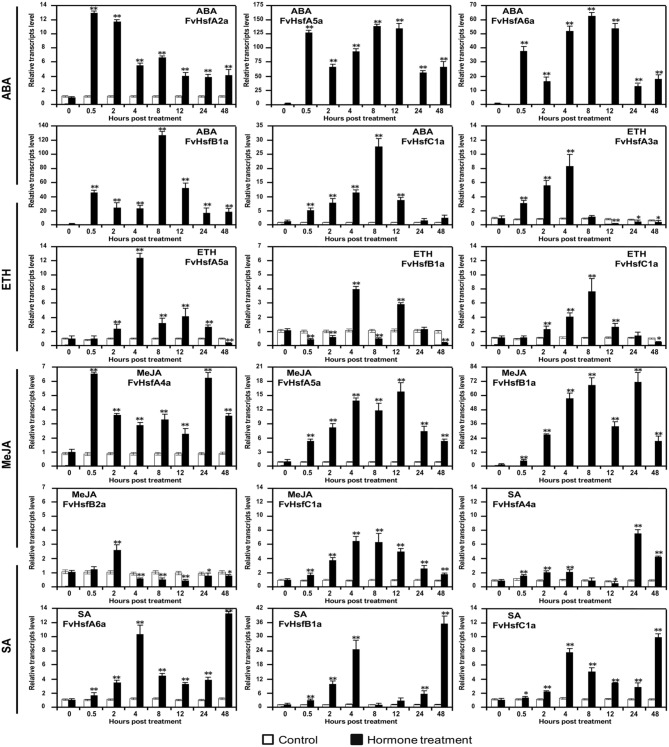
The expression of Hsf genes in the diploid woodland strawberry (F. vesca) in response to treatment with plant hormones. The expression levels of several representative FvHsf genes showed unusual patterns in response to ABA, SA, MeJA, and ethylene (Eth) treatments.
FvHsfA3a and FvHsfA4a were up-regulated 3- and 10-fold, respectively, during the 2–8 hpt period after 4°C treatment, whereas FvHsfA9a was up-regulated over 3-fold during the 24–48 hpt period of the same treatment (Figure 7A). FvHsfA3a was highly up-regulated 24–96 hpt after drought treatment, whereas FvHsfA4a, FvHsfB1a, and FvHsfC1a were initially up-regulated within 48 hpt and FvHsfA4a, FvHsfA5a, FvHsfA9a, FvHsfB1a, and FvHsfC1a were highly induced 96 hpt (Figure 7B). The FvHsfA4a, FvHsfA5a, FvHsfA9a, and FvHsfC1a transcripts returned to baseline or decreased when the plants were rewatered, but FvHsfB1a remained up-regulated 11-fold at this time point (Figure 7B). In addition, the FvHsfA2a transcript was up-regulated in the earlier stage of salt stress treatment before decreasing to its lowest level at 12 hpt (Figure 5A; Supplementary Figure S1). In contrast, the FvHsfA3a, FvHsfA5a, FvHsfA9a, and FvHsfB1a transcripts were primarily up-regulated at the middle or late stage of salt treatment. The level of the FvHsfC1a transcript first increased and then decreased during salt treatment (Figure 7C). After inoculation with Podosphaera aphanis, FvHsfB1a levels gradually increased during the earlier stage (24–96 hpi) and dramatically increased 120 hpi, whereas FvHsfA1b and FvHsfA3a were clearly down-regulated during the earlier stage (Figure 7D).
FvHsf expression in response to hormone treatments
Phytohormones such as abscisic acid (ABA), ethylene (Eth), salicylic acid (SA), and jasmonic acid (MeJA) have well-established roles in plant stress signaling networks (Kotak et al., 2007a; Clarke et al., 2009). To assess the potential roles of FvHsfs in phytohormone-mediated signal transduction, we examined the transcriptional abundance of 17 FvHsfs during ABA, Eth (ethephon), SA, and MeJA (Methyl jasmonate) treatments. Quantitative RT-PCR analysis revealed that ABA treatment causes the rapid (0.5 hpt) up-regulation of several FvHsf genes, including FvHsfA2a, FvHsfA5a, FvHsfA6a, FvHsfB1a, and FvHsfC1a. The levels of these transcripts remained higher (4- to 125-fold) throughout the entire treatment period (Figure 8). During Eth treatment, FvHsfA3a, FvHsfA5a, and FvHsfC1a were gradually up-regulated by 7.5- to 12-fold within 8 hpt, whereas FvHsfB1a was only up-regulated at 4 and 12 hpt. Treatment with MeJA also increased FvHsfA4a, FvHsfA5a, FvHsfB1a, and FvHsfC1a levels significantly (6- to 70-fold) throughout the entire treatment period, while FvHsfB2a was only up-regulated at 2 hpt before returning to a lower (~0.5-fold) abundance. During SA treatment, the FvHsfA4a, FvHsfA6a, FvHsfB1a, and FvHsfC1a transcripts exhibited a continuous increase: the transcript levels first peaked at 4 hpt and were again up-regulated during the 24–48 hpt time period (Figure 8). It is worth noting that FvHsfA4a, FvHsfA5a, FvHsfA6a, FvHsfB1a, and FvHsfC1a were clearly involved in the response to all four of the hormone treatments (Figures 5A, 8).
Discussion
F. vesca, a diploid woodland strawberry with a small and sequenced genome, is an excellent model for studying the counterparts of many important genes in the Rosaceae fruit crops (Shulaev et al., 2011; Darwish et al., 2015). An increasing amount of evidence has indicated that Hsf genes play essential roles in plant adaptations to various stress conditions (Shen et al., 2015; Xue et al., 2015). To explore Hsf functions in abiotic and biotic stress responses in the strawberry, this study identified and isolated Hsf genes in a diploid woodland strawberry in addition to analyzing the evolutionary relationships, gene structure, protein domains, subcellular localization, and expression profiles of these genes in response to abiotic or biotic stresses and hormone treatments.
The Hsf gene family is conserved in strawberry
The size of the strawberry Hsf gene family (17) is smaller than that of other plant species, such as Arabidopsis (21) (Scharf et al., 2012), rice (25) (Guo et al., 2008), apple (25) (Giorno et al., 2012), wheat (56) (Xue et al., 2014), and soybean (59) (Chung et al., 2013). The small size of the strawberry genome (Shulaev et al., 2011; Darwish et al., 2015) and lack of the large genome duplications seen in apple (Velasco et al., 2010), soybean (Schmutz et al., 2010), and wheat (Mayer et al., 2014) could explain the limited size of the FvHsf family (Figure 1A).
The comparative genomics approach structures genomes into syntenic blocks that exhibit conserved features (Ghiurcuta and Moret, 2014). This synteny analysis provided evolutionary and functional connections between genes in strawberry and Arabidopsis. Eleven strawberry Hsf genes were found to have syntenic relationships with Arabidopsis genes (Figure 1B), suggesting that most of the strawberry Hsf genes might have arisen before the divergence of the Arabidopsis and strawberry lineages. All of these FvHsf genes show close phylogenetic relationships with the corresponding AtHsf genes (Figure 1A), suggesting the potential for functional similarities. Structural characteristics such as intron-exon structures and protein domains might reflect the functional conservation or differentiation of the gene family. Fourteen of the seventeen FvHsf genes have only one phase zero intron while the others lack this intron (Figure 2), suggesting that the FvHsf gene family is conserved (de Souza et al., 1998). Besides, all 17 of the FvHsf proteins contain the necessary and/or specific protein domains (DBD, OD, NLS, NES, and AHA), which might be essential for functional conservation (Giorno et al., 2012). Fourteen of the FvHsf proteins retained the ability to localize in the nucleus, suggesting that most (14/17) of the FvHsf genes still maintain ancestral functional features (Figure 4).
FvHsf genes could play essential roles in strawberry adaptation to abiotic or biotic stresses
The function of plant Hsfs is to elicit the expression of genes encoding heat shock proteins (Hsps) or other stress-inducible genes (Schramm et al., 2006; Chan-Schaminet et al., 2009; Nishizawa-Yokoi et al., 2009) that are known to play a central role in protecting plants from heat or other stress conditions (Scharf et al., 2012). Recent genome-wide expression profile analyses of crop plants (rice, wheat, soybean, and apple) indicated that several Hsf genes are transcribed at high levels during heat, cold, salt, and drought stresses (Mittal et al., 2009; Giorno et al., 2012; Chung et al., 2013). In this study, 15 FvHsf genes showed distinct expression patterns during heat treatment (Figure 6), with most of these genes also were induced by other abiotic or biotic stresses and hormone stimuli (Figures 7, 8). It is notable that the subcellular localization of each FvHsf protein was different (Figure 4). All of our data suggest that the FvHsfs are involved in the response to almost all of the stress treatments, but these genes are highly specific in function.
FvHsfA2a, whose rice orthologs are OsHsfA2a/b/e, was found in the chromosomal region syntenic with AtHsfA2a (Figure 1). The characteristics of FvHsfA2a are highly consistent with the characteristics of OsHsfA2 and AtHsfA2a. For example, OsHsfA2a and AtHsfA2a have been reported as the most strongly induced Hsfs, accumulating to high levels in plants exposed to long-term heat stress (Schramm et al., 2006; Mittal et al., 2009). This finding is consistent with our results showing that FvHsfA2a was the most strongly induced of the 17 FvHsfs (~600-fold) in response to heat stress (Figure 6). In addition, AtHsfA2a was reported to play a broader role in the expression of multiple Hsps (Hsp70-5, Hsp18.1-Cl, and Hsp22-ER) (Schramm et al., 2006) and general stress-related non-chaperone encoding genes such as GOLS1 or APX2 (Nishizawa-Yokoi et al., 2009), while the FvHsfA2a transcript was clearly up-regulated in response to salt and ABA treatments (Figures 7C, 8). Lastly, while SlHsfA2a, a homolog of FvHsfA2a in tomato, reportedly localizes to the nucleus, interaction with SlHsfA1 is required for the efficient nuclear import of SlHsfA2a (Scharf et al., 1998). This finding is consistent with our results showing that FvHsfA2a-GFP fusion protein also localized to nucleus but could also be detected in the cytosol of Arabidopsis mesophyll protoplasts, which may result from the absence of the specific interaction factor (Figure 4).
FvHsfA3a has orthologs in apple (MdHsfA3a/b/c) and has a syntenic gene in Arabidopsis (AtHsfA3a) (Figure 1). MdHsfA3b/c are involved in maintaining the stress response when apple trees were exposed to prolonged periods of high temperature (Giorno et al., 2012). AtHsfA3a expression was regarded as part of the drought stress response (Schramm et al., 2008). SlHsfA3a, a homolog of FvHsfA3a in tomato, is a nuclear-localized Hsf that is induced by heat stress, high salinity, and drought, but not abscisic acid (Li et al., 2013). Our data are highly similar, with FvHsfA3a accumulating during heat, drought, salt, and ethephon treatments (Figures 6, 7B,C, 8) and FvHsfA3a-GFP fusion proteins clearly localized to the nucleus of protoplasts (Figure 4).
FvHsfA4a and FvHsfA5a are intriguing strawberry Hsfs. FvHsfA4a and FvHsfA5a were the earliest FvHsfs to diverge from the evolutionary branch, with similar results being found in the Hsf families of other species (Figure 1A). In addition, both FvHsfA4a and FvHsfA5a show significant localization to the cytosol of Arabidopsis mesophyll protoplasts (Figure 4). Notably, FvHsfA4a and FvHsfA5a were both distinctly up-regulated in response to abiotic stress (cold, drought, and salt) (Figure 7) and hormone treatments (ABA, Eth, MeJA, and SA) (Figure 8). The translocation of Hsf proteins from the cytosol to the nucleus is redox-dependent (Giesguth et al., 2015), and AtHsfA4a, an Arabidopsis ortholog of FvHsfA4a, has been shown to play a central role in the early sensing of H2O2 stress in Arabidopsis (Davletova et al., 2005). Thus, we speculate that the dramatically increased expression of FvHsfA4a and FvHsfA5a in response to a range of treatments (Figures 6–8) and the specific cellular localization of FvHsfA4a and FvHsfA5a (Figure 4) may have important implications for stress signaling.
It is notable that FvHsfA6a is an important member of the FvHsf family, with two syntenic AtHsfs in Arabidopsis: AtHsfA6b and AtHsfA7b (Figure 1B). Yoshida et al. confirmed that AtHsfA6a/b were involved in ABA-dependent signaling in response to water deficiency stress (Yoshida et al., 2010), and our data also showed that FvHsfA6a was highly expressed in response to ABA treatment (Figure 8). Recently, Xue et al. demonstrated that TaHsfA6f, a FvHsfA6a ortholog in Triticum aestivum, serves as a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Xue et al., 2015). This finding is consistent with FvHsfA6a showing high expression under heat treatment and highly specific nuclear localization (Figures 4, 6).
Undoubtedly, FvHsfA9a is a unique member of the FvHsf family. While FvHsfA9a is phylogenetically close to FvHsfA2a (Figure 1A), the expression characteristics of FvHsfA9a are entirely different from FvHsfA2a, which was primarily induced at the earlier stage of treatments (Figures 6, 7C, 8). In contrast, the current data identified FvHsfA9a as a late response factor in long-term heat, drought, salt, and cold stress treatments (Figures 6, 7). The Arabidopsis and Helianthus annuus homologs of FvHsfA9a function as seed-specific sHsp (small heat shock protein) regulators (Kotak et al., 2007; Prieto-Dapena et al., 2008) and are associated with ABA-mediated stress signaling and drought resistance (Schramm et al., 2008). Similarly, FvHsfA9a showed specific nuclear localization (Figure 4) and was only induced in response to ABA (Figure S2).
It is worth noting that FvHsfB1a was induced in response to all of the stress or hormone treatments, and was both instantly induced by all stimuli and also reached a high expression level during all of the treatments (Figures 6–8). Bharti et al. provided evidence that SlHsfB1, the tomato ortholog of FvHsfB1a, represents a novel type of coactivator, cooperating with class A Hsfs or other activators that control housekeeping gene expression during stress conditions (Bharti et al., 2004). Moreover, FvHsfB1a was highly induced at the late stage of powdery mildew infection (Figure 7) and accumulated to high levels during SA and MeJA treatments (Figure 8). Similarly, AtHsfB1a, the Arabidopsis homolog of FvHsfB1a, was reported as a crucial component of salicylic acid-mediated resistance (Kumar et al., 2009; Ikeda et al., 2011; Pick et al., 2012), and subsequent evidence also demonstrated that AtHsfB1a plays a pivotal role in primed defense gene activation and the pathogen-induced acquired immune response (Pick et al., 2012). Therefore, it is reasonable to speculate that FvHsfB1a is a key player in the acquired immune response to the biotrophic fungus Podosphaera aphanis.
In addition to stress, Hsfs have also been reported to play roles in plant growth and development (Begum et al., 2013). The detailed expression profiles of individual FvHsfs in six important organs were determined in this study (Figure 5B). Notably, several FvHsfs (FvHsfA2a, A3a, A4a, A5a, A6a, A9a, B1a, and C1a) exhibited organ-specific distribution and were clearly involved in stress responses (Figure 5A), suggesting that FvHsf members could be important in protecting growing or developing strawberry plants from heat damage.
Conclusion
In this study, we identified 17 FvHsf genes in a diploid strawberry by employing bioinformatics and publicly available data. The evolutionary relationship and structural feature analyses have aided in identifying the potential functions of individual strawberry Hsfs by retrieving clues from the well-investigated Arabidopsis. Numerous cis-acting elements were found in the FvHsf promoter sequences, suggesting that FvHsf gene expression is controlled by a complex regulatory regime. The gene expression profiles obtained during 42°C, 4°C, drought, salt, powdery mildew infection, and phytohormone treatments suggest that several strawberry Hsf genes (including FvHsfA2a, FvHsfA3a, FvHsfA4a, FvHsfA5a, FvHsfA6a, FvHsfA9a, FvHsfB1a, and FvHsfC1a) could play important roles in adaptation to environmental stresses. In addition, the distinct subcellular localization of strawberry Hsfs suggests the potential functional divergence of several strawberry Hsfs. Taken together, the present work may provide the basis for further studies to dissect FvHsf function in response to stress stimuli.
Author contributions
JF conceived and designed the experiments. YH, WW, and YTH performed the experiments. YH, KZ, WW, and JF analyzed the data. YH, YL, YG, and FZ contributed reagents/materials/analysis tools. YH and JF contributed to the writing of the manuscript. The authors thank Dr. Ke Duan of the Shanghai Academy of Agricultural Sciences for generously providing wild type Fragaria vesca plants.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31201657), the Fundamental Research Funds for the Central Universities (QN2013019), and the Shaanxi province science and technology research and development program (2014K02-02-02).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00736
Expression profiles of 17 FvHsf genes in response to abiotic and biotic stress treatments, as determined using semi quantitative RT-PCR.
Expression profiles of 17 FvHsf genes in response to phytohormone treatments, as determined using semi quantitative RT-PCR.
The primers used in this study.
The best matches for the protein motif sequences.
The syntenic relationships between strawberry and Arabidopsis Hsf genes.
References
- Almoguera C., Prieto-Dapena P., Díaz-Martín J., Espinosa J. M., Carranco R., Jordano J. (2009). The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol. 9:75. 10.1186/1471-2229-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum T., Reuter R., Schöffl F. (2013). Overexpression of AtHsfB4 induces specific effects on root development of Arabidopsis. Mech. Dev. 130, 54–60. 10.1016/j.mod.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Bharti K., Von Koskull-Döring P., Bharti S., Kumar P., Tintschl-Korbitzer A., Treuter E., et al. (2004). Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16, 1521–1535. 10.1105/tpc.019927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Schaminet K. Y., Baniwal S. K., Bublak D., Nover L., Scharf K. D. (2009). Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 284, 20848–20857. 10.1074/jbc.M109.007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E., Kim K. M., Lee J. H. (2013). Genome-wide analysis and molecular characterization of heat shock transcription factor family in Glycine max. J. Genet. Genomics 40, 127–135. 10.1016/j.jgg.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Clarke S. M., Cristescu S. M., Miersch O., Harren F. J. M., Wasternack C., Mur L. A. (2009). Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 182, 175–187. 10.1111/j.1469-8137.2008.02735.x [DOI] [PubMed] [Google Scholar]
- Darwish O., Shahan R., Liu Z. C., Slovin J. P., Alkharouf N. W. (2015). Re-annotation of the woodland strawberry (Fragaria vesca) genome. BMC Genomics 16:29. 10.1186/s12864-015-1221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D. J., Coutu J., et al. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17, 268–281. 10.1105/tpc.104.026971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza S. J., Long M., Kleln R. J., Roy S., Lin S., Gilbert W. (1998). Toward a resolution of the introns early/late debate: only phase zero introns are correlated with the structure of ancient proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 5094–5099. 10.1073/pnas.95.9.5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q. L., Ishii S., Sarai A. (1996). Binding site analysis of c-Myb: Screening of potential binding sites by using the mutation matrix derived from systematic binding affinity measurements. Nucleic Acids Res. 24, 766–774. 10.1093/nar/24.4.766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring P., Treuter E., Kistner C., Lyck R., Chen A., Nover L. (2000). The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12, 265–278. 10.1105/tpc.12.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiurcuta C. G., Moret B. M. E. (2014). Evaluating synteny for improved comparative studies. Bioinformatics 30, 9–18. 10.1093/bioinformatics/btu259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesguth M., Sahm A., Simon S., Dietz K. J. (2015). Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett. 589, 718–725. 10.1016/j.febslet.2015.01.039 [DOI] [PubMed] [Google Scholar]
- Giorno F., Guerriero G., Baric S., Mariani C. (2012). Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics 13:639. 10.1186/1471-2164-13-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. K., Wu J., Ji Q., Wang C., Luo L., Yuan Y., et al. (2008). Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genomics 35, 105–118. 10.1016/S1673-8527(08)60016-8 [DOI] [PubMed] [Google Scholar]
- Hahn A., Bublak D., Schleiff E., Scharf K. D. (2011). Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23, 741–755. 10.1105/tpc.110.076018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C. J., Bohm A. A., Nelson H. C. M. (1994). Crystal-structure of the DNA-binding domain of the heat-shock transcription factor. Science 263, 224–227. 10.1126/science.8284672 [DOI] [PubMed] [Google Scholar]
- Hedhly A., Hormaza J. I., Herrero M. (2009). Global warming and sexual plant reproduction. Trends Plant Sci. 14, 30–36. 10.1016/j.tplants.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Heerklotz D., Döring P., Bonzelius F., Winkelhaus S., Nover L. (2001). The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 21, 1759–1768. 10.1128/MCB.21.5.1759-1768.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. M., Kim D. W., Woo M. S., Jeong H. S., Son Y. S., Akhter S., et al. (2014). Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell Environ. 37, 1202–1222. 10.1111/pce.12228 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2011). Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254. 10.1104/pp.111.179036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Iu B., Singh J. (1996). Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 30, 679–684. 10.1007/BF00049344 [DOI] [PubMed] [Google Scholar]
- Kotak S., Port M., Ganguli A., Bicker F., Von Koskull-Döring P. (2004). Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 39, 98–112. 10.1111/j.1365-313X.2004.02111.x [DOI] [PubMed] [Google Scholar]
- Kotak S., Vierling E., Baumlein H., Von Koskull-Döring P. (2007). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19, 182–195. 10.1105/tpc.106.048165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Busch W., Birke H., Kemmerling B., Nürnberger T., Schöffl F. (2009). Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol. Plant 2, 152–165. 10.1093/mp/ssn095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Tang H. B., Wang X. Y., Paterson A. H. (2013). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158. 10.1093/nar/gks1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield O., Nelson H. C. M. (1999). A new use for the ‘wing’ of the ‘winged’ helix-turn-helix motif in the HSF-DNA cocrystal. Nat. Struct. Biol. 6, 464–470. 10.1038/8269 [DOI] [PubMed] [Google Scholar]
- Li Z., Zhang L., Wang A., Xu X., Li J. (2013). Ectopic overexpression of SlHsfA3, a heat stress transcription factor from tomato, confers increased thermotolerance and salt hypersensitivity in germination in transgenic Arabidopsis. PLoS ONE 8:e54880. 10.1371/journal.pone.0054880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Xie W. F., Zhang L., Valpuesta V., Ye Z. W., Gao Q. H., et al. (2014). Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J. Integr. Plant Biol. 56, 350–363. 10.1111/jipb.12150 [DOI] [PubMed] [Google Scholar]
- Lyck R., Harmening U., Höhfeld I., Treuter E., Scharf K. D., Nover L. (1997). Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta 202, 117–125. 10.1007/s004250050110 [DOI] [PubMed] [Google Scholar]
- Maughan T. L., Black B. L., Drost D. (2015). Critical temperature for sub-lethal cold injury of strawberry leaves. Sci. Hortic. 183, 8–12. 10.1016/j.scienta.2014.12.001 [DOI] [Google Scholar]
- Mayer K. F. X., Rogers J., Dolezel J., Pozniak C., Eversole K., Feuillet C., et al. (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 286. 10.1126/science.1251788 [DOI] [PubMed] [Google Scholar]
- Mittal D., Chakrabarti S., Sarkar A., Singh A., Grover A. (2009). Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 47, 785–795. 10.1016/j.plaphy.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Mittler R., Finka A., Goloubinoff P. (2012). How do plants feel the heat? Trends Biochem. Sci. 37, 118–125. 10.1016/j.tibs.2011.11.007 [DOI] [PubMed] [Google Scholar]
- Never L., Scharf K. D., Gagliardi D., Vergne P., Czarneckaverner E., Gurley W. B. (1996). The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones 1, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezhadahmadi A., Faruq G., Rashid K. (2015). The impact of drought stress on morphological and physiological parameters of three strawberry varieties in different growing conditions. Pak. J. Agric. Sci. 52, 79–92. [Google Scholar]
- Nishizawa-Yokoi A., Yoshida E., Yabuta Y., Shigeoka S. (2009). Analysis of the regulation of target genes by an Arabidopsis heat shock transcription factor, HsfA2. Biosci. Biotechnol. Biochem. 73, 890–895. 10.1271/bbb.80809 [DOI] [PubMed] [Google Scholar]
- Pick T., Jaskiewicz M., Peterhansel C., Conrath U. (2012). Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol. 159, 52–55. 10.1104/pp.111.191841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P., Castano R., Almoguera C., Jordano J. (2008). The ectopic overexpression of a seed-specific transcription factor, HaHSFA9, confers tolerance to severe dehydration in vegetative organs. Plant J. 54, 1004–1014. 10.1111/j.1365-313X.2008.03465.x [DOI] [PubMed] [Google Scholar]
- Raab T., Lopez-Raez J. A., Klein D., Caballero J. L., Moyano E., Schwab W., et al. (2006). FaQR, required for the biosynthesis of the strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, encodes an enone oxidoreductase. Plant Cell 18, 1023–1037. 10.1105/tpc.105.039784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wen L. P., Gao X. J., Jin C. J., Xue Y., Yao X. B. (2009). DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273. 10.1038/cr.2009.6 [DOI] [PubMed] [Google Scholar]
- Rombauts S., Dehais P., Van Montagu M., Rouze P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. 10.1093/nar/27.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K. D., Berberich T., Ebersberger I., Nover L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119. 10.1016/j.bbagrm.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Scharf K. D., Heider H., Hohfeld I., Lyck R., Schmidt E., Nover L. (1998). The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 18, 2240–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J. X., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Schramm F., Ganguli A., Kiehlmann E., Englich G., Walch D., Von Koskull-Doring P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 60, 759–772. 10.1007/s11103-005-5750-x [DOI] [PubMed] [Google Scholar]
- Schramm F., Larkindale J., Kiehlmann E., Ganguli A., Englich G., Vierling E., et al. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53, 264–274. 10.1111/j.1365-313X.2007.03334.x [DOI] [PubMed] [Google Scholar]
- Schwartz S., Meshorer E., Ast G. (2009). Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 16, 990–995. 10.1038/nsmb.1659 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Bevan M. (1998). The regulation of transcription factor activity in plants. Trends Plant Sci. 3, 378–383. 10.1016/S1360-1385(98)01302-8 [DOI] [Google Scholar]
- Shen Z., Yao J., Sun J., Chang L., Wang S., Ding M., et al. (2015). Populus euphratica HSF binds the promoter of WRKY1 to enhance salt tolerance. Plant Sci. 235, 89–100. 10.1016/j.plantsci.2015.03.006 [DOI] [PubMed] [Google Scholar]
- Shim D., Hwang J. U., Lee J., Lee S., Choi Y., An G., et al. (2009). Orthologs of the Class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21, 4031–4043. 10.1105/tpc.109.066902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V., Sargent D. J., Crowhurst R. N., Mockler T. C., Folkerts O., Delcher A. L., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109–116. 10.1038/ng.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42, 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Von Koskull-Doring P., Scharf K. D., Nover L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. 10.1016/j.tplants.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Xue G. P., Drenth J., Mcintyre C. L. (2015). TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 66, 1025–1039. 10.1093/jxb/eru462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G. P., Sadat S., Drenth J., Mcintyre C. L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557. 10.1093/jxb/ert399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. 10.1111/j.1365-313X.2009.04092.x [DOI] [PubMed] [Google Scholar]
- Zhang K., Han Y. T., Zhao F. L., Hu Y., Gao Y. R., Ma Y. F., et al. (2015). Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol. 15:164. 10.1186/s12870-015-0552-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profiles of 17 FvHsf genes in response to abiotic and biotic stress treatments, as determined using semi quantitative RT-PCR.
Expression profiles of 17 FvHsf genes in response to phytohormone treatments, as determined using semi quantitative RT-PCR.
The primers used in this study.
The best matches for the protein motif sequences.
The syntenic relationships between strawberry and Arabidopsis Hsf genes.