Abstract
Objective:
Single nucleotide polymorphisms (SNPs) in the promoter region of interleukin (IL)-10 gene, which codes for the anti-inflammatory cytokine IL-10, have been associated with its level of production in chronic periodontitis. The prevalence of promoter SNP genotypes is known in other populations with chronic periodontitis, while its association in the Indian population is not known. Hence, the present study was designed to investigate the prevalence of IL-10 promoter polymorphism in a racially defined group of Indians with severe chronic periodontitis as the Indian population is known to be genetically diverse.
Materials and Methods:
Genomic deoxyribonucleic acid was extracted from 46 nonsmoking patients with severe chronic periodontitis and 45 subjects with healthy periodontium. A SNP locus at −1087 of IL-10 was chosen, as this locus has been frequently associated with chronic periodontitis in other population. Genotyping was carried out using allele-specific polymerase chain reaction (AS-PCR), and the frequencies of genotype were analyzed between the groups.
Results:
The distribution of genotype and allele frequencies showed significant differences between the study groups. The prevalence of genotype AA alleles at −1087 locus of IL-10 was significantly higher in severe chronic periodontitis patients compared to the healthy controls (P = 0.05).
Conclusion:
The study has identified a positive association between the occurrence of AA allele at −1087 locus of IL-10 gene and severe chronic periodontitis in nonsmoking patients.
Keywords: Interleukin-10 alleles in periodontitis, interleukin-10 polymorphism in periodontitis, interleukin-10 polymorphism, promoter polymorphism in periodontitis, single nucleotide polymorphism in periodontitis
INTRODUCTION
Chronic periodontitis (CP) is an inflammatory disease of periodontium, which develops as a response to toxic factors secreted by periodontal pathogens. CP is a progressive condition that leads to gradual recession of periodontium accompanied by loss of supporting bone with tooth mobility as a consequence.[1] Nearly 10% of the adult population are affected by severe forms of CP. The etiology is understood to be complex and multifactorial and are attributed to risk factors like plaque microflora, tobacco smoking,[1] systemic diseases like diabetes[1] and genetic factors.[2] Deciphering the interplay between these factors is thus essential to understand the pathogenesis of the disease. Initiation and progression of severe CP begins with toxic factors secreted by periodontal pathogens that promote the inflammatory process by triggering the production of inflammatory cytokines. Cytokines are usually produced transiently but in a regulated manner in varying concentrations by large number of cells such as T-cells, B-cells, macrophages, and monocytes that have different range of activity in order to effectively orchestrate the inflammatory response to injury.[3] Hence, dysregulation in the production of cytokines may be expected to cause a subdued or accentuated response leading to untoward tissue destruction and disease progression.
Association of single nucleotide polymorphism (SNPs) with periodontal disease was first reported by Kornman et al. in 1997[4] where the authors found association of SNPs in interleukin (IL)-1A (−889 in linkage with +4845), IL-1B (−511in linkage with −31), IL-1B (+3954) and IL-1RN (variable number tandem repeat in linkage with +2018) with severe CP. Several genetic polymorphism studies on IL-1, tumor necrosis factor-α, IL-4, IL-6, IL-10, FcγRIIa, FcγRIIIa, FcγRIIIb, Vitamin D receptor, CD14, TLR2, and TLR4 genes have since been reported in different populations with periodontal disease. Of the above genes, IL-10 synthesizes an inflammatory cytokine IL-10, which is produced by monocytes, Th2 cells, regulatory T-cells, and B-cells.[5] The presence of IL-10 in periodontitis was first reported by Gemmel et al.[6] and Yamazaki et al.,[7] who investigated the level of IL-10 mRNA by real time-polymerase chain reaction and found higher levels of IL-10 mRNA in gingival biopsies of severe CP patients relative to peripheral blood mononuclear cells.
Single nucleotide polymorphisms in the promoter region of IL-10 gene have been identified at positions −1087,−592, −819 from the transcriptional start site, and have been associated with a number of inflammatory diseases like juvenile rheumatoid arthritis,[8] systemic lupus erythematosus,[9,10] inflammatory bowel disease,[11] bronchial asthma,[12] and CP. In case of CP, several independent studies have identified association of the SNPs with extent and/or severity of CP, albeit at varying frequencies in patients from diverse racial background [Table 1].
Table 1.
Incidence of association of −1087, −819, and −592 SNPs in promoter of IL-10 gene with CP

Although the genetic background of Indian population has been established to be unique from that of HapMap population,[23] and that the genetic background is known to be diverse among the population in different geographical locations within India, the status of IL-10 promoter SNPs in Indian patients with CP remains yet to be investigated. In order to address this issue, the present study was designed as a pilot explorative study by including, (a) Racially defined Tamil speaking Dravidians so as to eliminate confounding effect that may arise due to genetic diversity, and (b) IL-10 SNP at −1087 alone as it has been shown to be consistently associated with chronic periodontitis in the European patients [Table 1].[13,19,20]
MATERIALS AND METHODS
Subject selection
A total of 91 subjects were included in the study after obtaining informed consent from the patients. The study was approved by the Ethical Committee and Scientific Committee of the Institutional Review Board. The study group included 46 severe CP patients as the test group (Group A) and 45 periodontally healthy patients as the control (Group B). Patients from both genders in the age group 30–50 years were included when presented with clinical attachment loss ≥5 mm, probing pocket depth (PPD) ≥5 mm involving ≥30% sites. Patients with above clinical presentation but with systemic diseases were excluded. Periodontally healthy subjects with no signs of periodontal disease and probing sulcus depth of >3 mm were selected for the control group. Both the test group and control group of patients belonged to Tamil speaking Dravidian race.
Clinical evaluation of the subjects
The evaluation of each subject was done, based on their demographic details, ethnicity, mother tongue, and racial background. A brief medical, dental history as reported by the patient and clinical parameters were recorded. Diagnosis and classification of periodontal disease status were established based on clinical parameters which included PPD and clinical attachment level (CAL) measured at six sites around each tooth using a University of North Carolina probe. The probe was directed to the long axis of the tooth. The indices taken for the study included standard dental evaluation procedures namely, plaque index (PlI), oral hygiene index-simplified (OHI-S) and modified sulcular bleeding index (mSBI). All clinical recordings were performed by a single examiner. Intraexaminer calibration was achieved by two examinations of ten patients, 24 h before recruiting the subjects into the study. Calibration was accepted if measurements at baseline and after 24 h were similar to 1 mm at the 95% level.
Sample collection and deoxyribonucleic acid extraction
One milliliter of venous blood was collected by venipuncture from each patient in ethylenediaminetetraacetic acid (EDTA) coated tubes and stored at −80°C. At the time of deoxyribonucleic acid (DNA) extraction, blood samples were thawed and centrifuged at 500 rpm for 3 min at room temperature to pellet blood cells. Pelleted cells were washed thrice with red cell lysis buffer (containing 0.05% of saponin in 1X phosphate buffered saline (PBS), pH 7.5) to remove red blood cells from the whole blood. After the final wash, white blood cells appeared as a clear buffy coat to which 1 ml of cell lysis buffer (containing 0.1% sodium dodecyl sulfate, 25 mM EDTA, 75 µg/100 µl proteinase-K, and 200 mM Tris-Cl at pH 8 [Sigma-Aldrich, St. Louis, MO, USA]) was added and incubated at 57°C for 12 h with intermittent agitation. Cell lysates were then treated with 200 µl of 100 mM Ammonium Acetate, mixed and incubated at room temperature for 15 min. The samples were then centrifuged at 12,000 rpm for 15 min to precipitate protein fraction in cell lysate. Following this step, one-sixth volume of isopropanol was added to all the samples, mixed by vortexing and centrifuged at 12,500 rpm for 15 min at room temperature to precipitate the genomic DNA.
Allele specific polymerase chain reaction
The forward primer of allele-specific polymerase chain reaction (PCR), flanking −1087 region in IL-10 promoter was designed as described earlier,[24] and is as follows:
Interleukin-10F1: CTA AGG CTT CTT TGG GAA (−1087A specific)
Interleukin-10F2: CTA AGG CTT CTT TGG GAG (−1087G specific).
Each of the forward primer specifically binds only to its respective allele.
The reverse primer was designed as a universal primer, the sequence of which is as follows:
Interleukin-10R: AGG CAC ATG TTT CCA CCT CTT CAG (common to both −1087A and −1087G alleles).
The primer sequences were synthesized at Genorime Facility, Chennai.
Polymerase chain reaction was performed under the following conditions: Initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 45 s, primer annealing at 60°C for 60 s, primer extension at 72°C for 60 s, with a final extension at 72°C for 5 min.
Statistical analysis
The data obtained were formulated and statistically analyzed using SPSS v17 software (IBM corporation). The basic characteristics of the study population like age and gender were analyzed using independent sample t-test. The mean of clinical parameters like oral hygiene index, PPD, CAL, PlI, and mSBI were calculated and the intragroup and intergroup comparisons were derived using Independent sample t-test. The Chi-squared test was applied to examine the differences in genotype distribution, allele frequency between test and control groups. Deviation from Hardy–Weinberg equilibrium was assessed by goodness-of-fit between the observed and expected numbers using Chi-square test with 1 degree of freedom. The comparison of clinical parameters with the genotype was performed within the groups and between the groups using one-way analysis of variance (ANOVA) analysis and the P value was obtained thereafter.
RESULTS
The study was undertaken to investigate the role of IL-10 at −1087 loci in patients with severe CP relative to age, gender and race-matched controls without periodontitis. Power calculations performed initially showed that the sample size required for ascertaining the significance of association of periodontal disease and genetic polymorphism with an alpha error of 1% and 90% power to be 41 for both severe CP and control groups. Based on this criterion, a total of 91 subjects, 46 with severe CP group, and 45 healthy controls were included in the study.
Age and gender distribution between severe chronic periodontitis and control groups
The mean age was 41.89 years in severe CP and 43.13 years in healthy controls. Independent t-test with equal variance between the two groups revealed a P = 0.094 which was not statistically significant and hence was considered to depict age-matched individuals between the two groups. A total of 47 male subjects and 44 female subjects were present in both groups. Both Pearson Chi-square (χ2 = 0.271) and P = 0.378 values were not significant between the groups that showed gender match between both severe CP group and healthy controls [Table 2].
Table 2.
Age and gender distribution between severe CP and healthy control groups

Clinical presentation in severe chronic periodontitis and healthy control groups
The mean of scores of OHI-S, PlI, and mSBI were significantly higher in severe CP group relative to healthy controls group (P < 0.001). The mean of PPD and CAL were also several fold higher in subjects with severe CP than the healthy controls (P < 0.001) [Table 3].
Table 3.
Comparison of mean values of the clinical parameters among the severe CP and healthy control groups

Distribution of genotypes in severe chronic periodontitis and healthy control groups
The presence of genotype GG at −1087 locus of IL-10 gene produces significantly higher amount of IL-10 cytokine than IL-10 gene with AA genotype. Hence, in order to ascertain the occurrence of the genotype in the study samples, PCR was performed by combining either IL-10F1 and IL-10R to determine A allele or IL-10F2 and IL-10R to determine G allele on DNA samples extracted from severe CP and control groups. A representative gel image of the samples along with positive controls is shown in Figure 1. Analysis of the distribution of genotypes among the study groups showed a higher percent occurrence of AA genotype in severe CP patients and GG genotype in healthy controls group subjects. The distribution of heterozygous AG genotype was comparable in both groups. Pearson χ2 = 6.285 (P = 0.043) and χ2 = 5.8855 (P = 0.0152) were obtained when either the three genotypes or alleles were compared among the CP and control groups [Table 4]. Hardy–Weinberg equilibrium analysis of the genotype distribution among the CP and control groups showed that the probability of differences between the observed and expected values due to chance to be very low (χ2 = 9.011, P < 0.0026 with 1 degree of freedom) [Table 5].
Figure 1.

Representative gel image of allele-specific polymerase chain reaction of severe CP group and healthy control group samples
Table 4.
Percentage distribution of genotypes and allele frequencies in severe CP and healthy controls groups, along with logistic regression analysis of IL-10 AG and AA genotypes with reference to GG, and A allele with reference to G

Table 5.
Hardy-Weinberg equilibrium for genotypes

Distribution of allele frequency in severe chronic periodontitis group and healthy controls group
To enumerate G or A alleles that occur in the genotype at −1087 locus of IL-10 gene in the study groups, the data were analyzed by Pearson Chi-square test. Data analysis showed higher distribution of G allele in healthy controls (n = 26%, 66.66%) and A allele in severe CP (n = 79%, 55.24%), which indicated a clear association of low cytokine producing allele A with severe CP. Since the A alleles occurred in at higher frequency in patients with severe CP, we next performed logistic regression analysis of the two genotypes - AG and AA with GG as reference, which showed a significance association of AA genotype with severe CP (odds ratio [OR] = 10.07%, 95% confidence interval [CI] = 1.1860–85.57). Similar analysis for A allele with G as reference showed significant association of A allele with severe CP (OR = 2.46%, 95% CI = 1.17–5.18) [Table 4].
Comparison of genotypes in severe chronic periodontitis and healthy control groups with clinical parameters by one-way analysis of variance
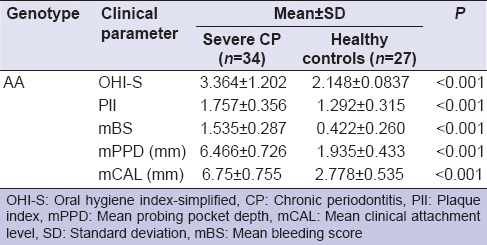
Since the allele frequency of A was higher in severe CP group, we next analyzed to understand the association of AA genotype with each of the clinical parameter. One-way ANOVA analysis showed highly significant association of AA genotype with all five clinical parameters namely, OHI-S, PlI, mBS, mean probing pocket depth, and mean clinical attachment level (P ≤ 0.001) [Table 6].
Table 6.
Association of genotype AA with clinical parameters between the severe CP and healthy control groups
DISCUSSION
Genetically predetermined capability of an individual to respond to microbial and environmental factors has been shown to be associated with the susceptibility for CP. In particular, polymorphisms in genes encoding molecules of the host defense system, such as cytokines, have been targeted as potential genetic markers. IL-10 is one such anti-inflammatory cytokine, which plays a role in periodontitis by down-regulating the production of monocyte-derived pro-inflammatory cytokines and stimulating protective antibody production.[14,15] Three SNPs at positions −1087,−819, and −592 from transcriptional start site of IL-10 have been associated with several inflammatory diseases including CP, and genotype at position −1087 has been associated with production of IL-10 cytokine in in vitro studies. In order to ascertain the prevalence of genotype at −1087, we analyzed 46 subjects with severe CP along with 45 subjects with healthy periodontium to understand the prevalence of SNPs in the promoter of IL-10. The subjects in both the groups were matched for age, gender and race. To eliminate possible confounding effects due to tobacco and other systemic diseases, only nonsmokers with no known systemic diseases were analyzed. The polymorphism of IL-10 at loci −1087 was assessed by allele-specific polymerase chain reaction technique (AS-PCR), which has been successfully used in earlier studies.[24] AS-PCR on the study samples indicated a positive association of −1087(A/G) SNP with severe CP, which was determined based on: (1) Higher percent prevalence of lower IL-10 producing genotype AA in severe CP patients, and (2) higher percent occurrence of increased IL-10 producing genotype GG genotype in periodontally healthy individuals. A significant association of clinical symptoms such as increase in gingival inflammation, probing sulcus depth, and clinical attachment loss was observed in patients carrying AA genotype relative to other genotype (AG or GG) carriers. The finding may be considered as clinically significant as the sample analysis was done on a racially defined group, Tamil speaking Dravidians from the Southern region of India. Racial segregation of the samples was done to avoid confounding effects that may arise due to genetic diversity as the Indian population is known to be genetically diverse.[23] Hardy–Weinberg equilibrium analysis confirmed the distribution of genotypes to be in harmony (χ2 = 9.011, P < 0.002) among the tested samples, which clearly suggested the occurrence of allele and genotype frequencies to be constant from generation to generation in an infinitely large interbreeding population. The higher occurrence of AA in severe CP patients and GG genotype in age and race-matched healthy individuals together advocates for a protective effect of GG allele in the analyzed samples, which is in agreement with earlier studies.[6,24]
While the findings of the present study strongly support for the association of AA genotype at −1087 loci with CP, studies elsewhere excepting two on Swedish population have observed otherwise. Most of them have identified the positive association of SNPs at −819 and −592 loci with CP. Hence, we believe that the inclusion of −819 and −592 loci as well in the present analysis would have: (a) Provided a comparative prevalence pattern relative to other population, and (b) increased the significance of clinical association of IL-10 SNPs with severe CP.
It is noteworthy to mention that the genetic status of IL-10 cannot be considered as the sole determinant in the development of CP, as the etiology of CP is known to be multifactorial in nature. In our own study, we observed the low IL-10 producing genotype AA in 60% of subjects from the healthy control group as against 73.9% from severe CP group. Despite of having the AA genotype, the age-matched subjects in the control group did not develop periodontitis. This observation reiterates the genetic component as a risk factor, that is, in the presence of good oral hygiene measures the role of AA genotype remains insignificant. However, in the presence of precipitating factors the periodontium of AA genotype carriers suffer a dual insult, (1) Inflammatory reaction triggered by microbial toxins, and (2) lower production of IL-10 that may not be proportionate enough to extent of periodontitis, which accelerates the progression of periodontitis. It is also possible that the presence of polymorphisms in other ILs besides IL-10 SNP may also be essential for the progression of severe CP. For example, polymorphism in IL-1 and levels of IL-1, IL-8, and IL-17 in gingival crevicular fluid have been shown to be associated with severity of the periodontal condition.[25,26,27]
To best of our knowledge, this is the first case-control study to have demonstrated a positive association between the −1087 IL-10 promoter polymorphism and severe CP in the Indian population. Although only a small representative population was investigated, the statistically significant association of −1087 SNP highlights for the requirement of a large scale case-controlled prevalence study by including age and gender-matched patients with severe CP from different races across India. The findings from these studies may then be effectively used to identify subjects with a higher risk of developing severe CP, so that appropriate preventive measures may be followed.
CONCLUSION
This is the first case-control study from India to investigate and identify a higher incidence of the decreased IL-10 producing genotype AA at −1087 loci in nonsmokers with severe CP, which indicated A allele at −1087 loci as a risk factor for the development of severe CP in subjects with poor oral hygiene.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Kobayashi T, Yoshie H. Host responses in the link between periodontitis and rheumatoid arthritis. Curr Oral Health Rep. 2015;2:1–8. doi: 10.1007/s40496-014-0039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michalowicz BS, Diehl SR, Gunsolley JC, Sparks BS, Brooks CN, Koertge TE, et al. Evidence of a substantial genetic basis for risk of adult periodontitis. J Periodontol. 2000;71:1699–707. doi: 10.1902/jop.2000.71.11.1699. [DOI] [PubMed] [Google Scholar]
- 3.Seymour GJ, Taylor JJ. Shouts and whispers: An introduction to immunoregulation in periodontal disease. Periodontol 2000. 2004;35:9–13. doi: 10.1111/j.0906-6713.2004.003555.x. [DOI] [PubMed] [Google Scholar]
- 4.Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW, et al. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;24:72–7. doi: 10.1111/j.1600-051x.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 5.Gonzales JR, Michel J, Diete A, Herrmann JM, Bödeker RH, Meyle J. Analysis of genetic polymorphisms at the interleukin-10 loci in aggressive and chronic periodontitis. J Clin Periodontol. 2002;29:816–22. doi: 10.1034/j.1600-051x.2002.290905.x. [DOI] [PubMed] [Google Scholar]
- 6.Gemmell E, Marshall RI, Seymour GJ. Cytokines and prostaglandins in immune homeostasis and tissue destruction in periodontal disease. Periodontol 2000. 1997;14:112–43. doi: 10.1111/j.1600-0757.1997.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 7.Yamazaki K, Tabeta K, Nakajima T, Ohsawa Y, Ueki K, Itoh H, et al. Interleukin-10 gene promoter polymorphism in Japanese patients with adult and early-onset periodontitis. J Clin Periodontol. 2001;28:828–32. doi: 10.1034/j.1600-051x.2001.028009828.x. [DOI] [PubMed] [Google Scholar]
- 8.Crawley E, Kay R, Sillibourne J, Patel P, Hutchinson I, Woo P. Polymorphic haplotypes of the interleukin-10 5' flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum. 1999;42:1101–8. doi: 10.1002/1529-0131(199906)42:6<1101::AID-ANR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 9.Pacheco AG, Cardoso CC, Moraes MO. IFNG +874T/A, IL10-1082G/A and TNF-308G/A polymorphisms in association with tuberculosis susceptibility: A meta-analysis study. Hum Genet. 2008;123:477–84. doi: 10.1007/s00439-008-0497-5. [DOI] [PubMed] [Google Scholar]
- 10.Pyo CW, Hur SS, Kim YK, Choi HB, Hong YS, Kim DW, et al. Polymorphisms of IL-1B, IL-1RN, IL-2, IL-4, IL-6, IL-10, and IFN-gamma genes in the Korean population. Hum Immunol. 2003;64:979–89. doi: 10.1016/s0198-8859(03)00173-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhu H, Lei X, Liu Q, Wang Y. Interleukin-10-1082A/G polymorphism and inflammatory bowel disease susceptibility: A meta-analysis based on 17,585 subjects. Cytokine. 2013;61:146–53. doi: 10.1016/j.cyto.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Hyun MH, Lee CH, Kang MH, Park BK, Lee YH. Interleukin-10 promoter gene polymorphisms and susceptibility to asthma: A meta-analysis. PLoS One. 2013:8–e53758. doi: 10.1371/journal.pone.0053758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berglundh T, Donati M, Hahn-Zoric M, Hanson LA, Padyukov L. Association of the-1087 IL 10 gene polymorphism with severe chronic periodontitis in Swedish Caucasians. J Clin Periodontol. 2003;30:249–54. doi: 10.1034/j.1600-051x.2003.10274.x. [DOI] [PubMed] [Google Scholar]
- 14.Scarel-Caminaga RM, Trevilatto PC, Souza AP, Brito RB, Camargo LE, Line SR. Interleukin 10 gene promoter polymorphisms are associated with chronic periodontitis. J Clin Periodontol. 2004;31:443–8. doi: 10.1111/j.1600-051X.2004.00500.x. [DOI] [PubMed] [Google Scholar]
- 15.Babel N, Cherepnev G, Babel D, Tropmann A, Hammer M, Volk HD, et al. Analysis of tumor necrosis factor-alpha, transforming growth factor-beta, interleukin-10, IL-6, and interferon-gamma gene polymorphisms in patients with chronic periodontitis. J Periodontol. 2006;77:1978–83. doi: 10.1902/jop.2006.050315. [DOI] [PubMed] [Google Scholar]
- 16.Sumer AP, Kara N, Keles GC, Gunes S, Koprulu H, Bagci H. Association of interleukin-10 gene polymorphisms with severe generalized chronic periodontitis. J Periodontol. 2007;78:493–7. doi: 10.1902/jop.2007.060309. [DOI] [PubMed] [Google Scholar]
- 17.Mellati E, Arab HR, Tavakkol-Afshari J, Ebadian AR, Radvar M. Analysis of-1082 IL-10 gene polymorphism in Iranian patients with generalized aggressive periodontitis. Med Sci Monit. 2007;13:CR510–4. [PubMed] [Google Scholar]
- 18.Claudino M, Trombone AP, Cardoso CR, Ferreira SB, Jr, Martins W, Jr, Assis GF, et al. The broad effects of the functional IL-10 promoter-592 polymorphism: Modulation of IL-10, TIMP-3, and OPG expression and their association with periodontal disease outcome. J Leukoc Biol. 2008;84:1565–73. doi: 10.1189/jlb.0308184. [DOI] [PubMed] [Google Scholar]
- 19.Donati M, Liljenberg B, Padyukov L, Berglundh T. Local expression of interleukin-10 and mCD14 in relation to the-1087 IL-10 and-159 CD14 gene polymorphisms in chronic periodontitis. J Periodontol. 2008;79:517–24. doi: 10.1902/jop.2008.070299. [DOI] [PubMed] [Google Scholar]
- 20.Reichert S, Machulla HK, Klapproth J, Zimmermann U, Reichert Y, Gläser CH, et al. The interleukin-10 promoter haplotype ATA is a putative risk factor for aggressive periodontitis. J Periodontal Res. 2008;43:40–7. doi: 10.1111/j.1600-0765.2007.00992.x. [DOI] [PubMed] [Google Scholar]
- 21.Hu KF, Huang KC, Ho YP, Lin YC, Ho KY, Wu YM, et al. Interleukin-10 (-592 C/A) and interleukin-12B (+16974 A/C) gene polymorphisms and the interleukin-10 ATA haplotype are associated with periodontitis in a Taiwanese population. J Periodontal Res. 2009;44:378–85. doi: 10.1111/j.1600-0765.2008.01116.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhong Q, Ding C, Wang M, Sun Y, Xu Y. Interleukin-10 gene polymorphisms and chronic/aggressive periodontitis susceptibility: A meta-analysis based on 14 case-control studies. Cytokine. 2012;60:47–54. doi: 10.1016/j.cyto.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Indian Genome Variation Consortium. Genetic landscape of the people of India: A canvas for disease gene exploration. J Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- 24.Koss K, Satsangi J, Fanning GC, Welsh KI, Jewell DP. Cytokine (TNF alpha, LT alpha and IL-10) polymorphisms in inflammatory bowel diseases and normal controls: Differential effects on production and allele frequencies. Genes Immun. 2000;1:185–90. doi: 10.1038/sj.gene.6363657. [DOI] [PubMed] [Google Scholar]
- 25.Kayar NA, Alptekin NO, Haliloglu S. Interleukin-1 receptor antagonist levels in gingival crevicular fluid and serum in nonsmoking women with preterm low birth weight and intrauterine growth retardation. Eur J Dent. 2015;9:109–16. doi: 10.4103/1305-7456.149655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archana PM, Salman AA, Kumar TS, Saraswathi PK, Panishankar KH, Kumarasamy P. Association between interleukin-1 gene polymorphism and severity of chronic periodontitis in a south Indian population group. J Indian Soc Periodontol. 2012;16:174–8. doi: 10.4103/0972-124X.99258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erdemir EO, Hendek MK, Keceli HG, Apan TZ. Crevicular fluid levels of interleukin-8, interleukin-17 and soluble intercellular adhesion molecule-1 after regenerative periodontal therapy. Eur J Dent. 2015;9:60–5. doi: 10.4103/1305-7456.149644. [DOI] [PMC free article] [PubMed] [Google Scholar]


