Figure 1. Overview of the methods of identifying species-species and species-location interactions.
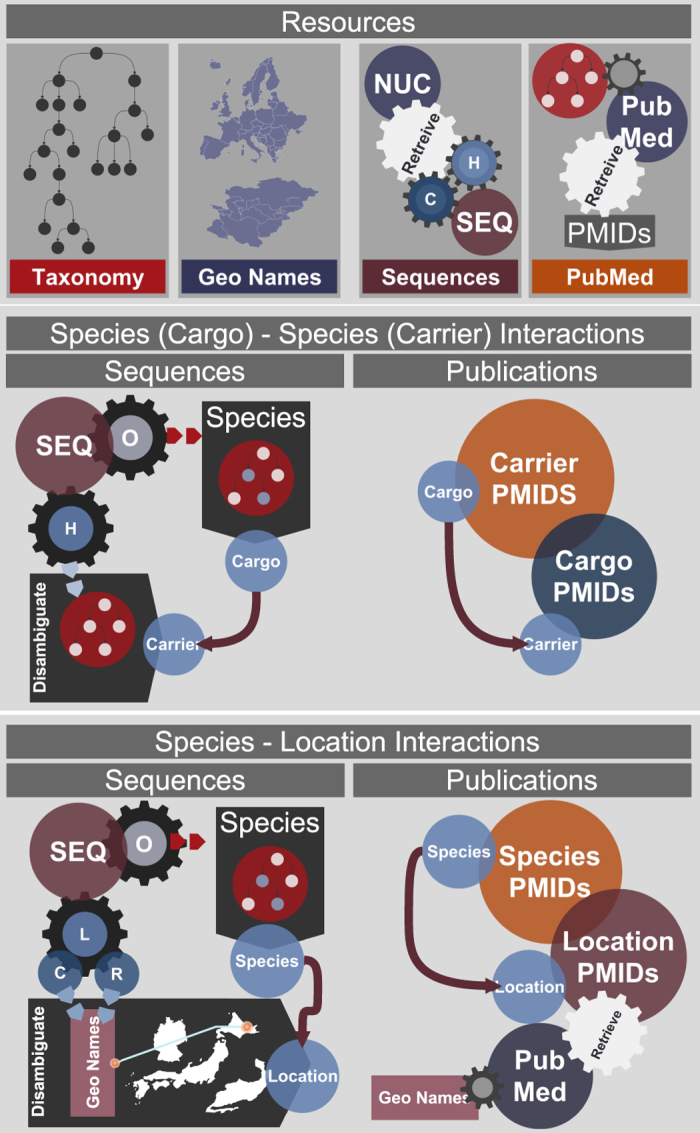
The first panel lists the resources used in a colour coded fashion. H refers to host and C to country tags in the sequence metadata. PMID is the PubMed unique identifier used in retrieving papers. The second panel explains the method of interrogating the evidence bases to extract species (cargo)-species (carrier) interactions. Species of sequenced organism (i.e., cargo) is first identified using the taxonomy tree, then the host tag in the sequence metadata is disambiguated using the taxonomic tree to identify the carrier species. Lists of PMIDs obtained for cargo and carrier species are intersected to provide additional evidence for the interactions extracted from the sequence metadata and to identify new relationships between cargo and carrier species discovered from the sequence metadata. The third panel illustrates the method of extracting species-location interactions from the evidence-base. First sequenced organisms and location information are extracted from sequence metadata. The species of sequenced organisms is then identified using the taxonomic tree. The location data (L) is split into country (C) and region (R) strings. Both are then disambiguated using the data gathered from GeoNames to obtain the country and region where the species was found. Geonames is also used to interrogate PubMed for papers about each country and region in the database. These are then intersected with species publications, the shared set is used as evidence for the species being found in a given location.
