Abstract
Background
MicroRNAs (miRNAs) are small non-coding RNAs that regulate the gene expression of target mRNAs involved in plant growth, development, and abiotic stress and pathogen responses. Previous studies have reported miRNAs in Populus that respond to abiotic stresses, such as cold, heat, drought, flooding, high salt and mechanical stress. However, little is known about the regulatory roles of these molecules in the Populus response to the stress of foliar rust fungal infection. Here, we identified the miRNA profiles of Populus after inoculation with Melampsora larici-populina using high-throughput sequencing and bioinformatics analysis. Quantitative real-time PCR (qRT-PCR) was used to validate the expression levels of 10 miRNAs.
Results
A total of 90 known miRNAs belonging to 42 families and 378 novel miRNAs were identified from three small-RNA libraries of Populus szechuanica infected with M. larici-populina isolates Sb052 and Th053 and a control. Comparative analysis revealed that the expression of 38 known miRNAs and 92 novel miRNAs in P. szechuanica after infection with different rust fungus isolates showed significant differences, and more miRNAs were suppressed during rust infection. Among the differentially expressed miRNAs, 7 known and 20 novel miRNAs were relevant to the rust fungus infection, and according to KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathway analysis, these miRNAs primarily regulate genes encoding disease-resistance proteins, serine/threonine protein kinases, transcription factors, and related proteins. QRT-PCR analysis indicated that most miRNAs were up-regulated in the Sb052 library and down-regulated in the Th053 library at 48 h post-inoculation (hpi).
Conclusions
These results demonstrate that the expression of miRNAs was altered in poplar under stress associated with M. larici-populina infection, and different temporal dynamics were observed in incompatible and compatible libraries. These findings suggest important roles for miRNA regulation in Populus upon infection with foliar rust fungus.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1891-8) contains supplementary material, which is available to authorized users.
Keywords: Populus szechuanica, MicroRNA, High-throughput sequencing, Melampsora larici-populina
Background
MiRNAs, small endogenous non-coding RNAs approximately 21–24 nucleotides (nt) in length, play an important role in regulating gene expression at the post-transcriptional level [1]. A large number of miRNAs have recently been identified in plants, and numerous miRNAs have been entered into the miRBase. miRNAs are involved in regulating growth, development, hormone balance, floral morphogenesis, reproductive performance, and biotic and abiotic stress responses. Upon nutritional deficiency, miRNAs regulate the metabolic balance of phosphorus, sulphur and copper in plants [2–5]. Under environmental stresses, including drought, flooding, salt, cold and heat, stress-regulated miRNAs in plants confer resistance to the extreme conditions [6]. In addition, the expression of miRNAs can alter in plants in response to biotic stresses, such as fungi, bacteria, viruses, or insects [7–16].
The genome of P. trichocarpa is small, and these plants reach reproductive maturity in a relatively short time [17, 18]. Thus, Populus is a useful forest species model for genetic and ecological research. In recent years, studies on Populus miRNAs have increased [19–21], particularly the expression of miRNAs and the function of the targets in response to abiotic stresses, including cold, heat, drought, flooding, high salt and mechanical stress [22–29]. A larger number of serious diseases occur in poplar, particularly foliar rust disease induced through infections with the rust fungus Melampsora spp., which markedly affects the growth of seeding and saplings. However, few studies have focused on miRNA expression profiling in the response of poplars to biotic stress, particularly pathogen stress [30, 31].
To understand the roles of miRNAs in the response of poplars to pathogenic fungi, we constructed three libraries from uninfected P. szechuanica leaves (control) and leaves infected with avirulent and virulent isolates of M. larici-populina, detected the expression of miRNAs through high-throughput sequencing and analysed the target genes. QRT-PCR was used to analyse the expression of ten miRNAs in plants inoculated for different times. The results of the present study help elucidate the regulatory mechanisms of Populus in response to foliar rust fungal infections.
Results
Analysis of miRNA sequences
Using Solexa sequencing, we constructed three libraries from uninoculated P. szechuanica leaves (control) and leaves inoculated with avirulent (Sb052) and virulent (Th053) isolates of M. larici-populina. A total of 17,801,357, 19,602,945 and 17,962,927 raw reads were obtained from Sb052, Th053 and control libraries, respectively (Table 1). After eliminating low-quality reads and impurities, 17,754,456, 19,544,426 and 17,918,326 high-quality reads were obtained from the three libraries. After discarding 3’ adapter deletions, insertion deletions, 5’ adapter contaminants, poly-A sequences and sequences less than 18 nt from the high-quality reads, 17,557,485, 19,398,402 and 17,764,475 clean reads were used for further analysis. The proportions of clean reads were 98.89, 99.25 and 99.14 % of the total reads obtained from the three libraries, respectively.
Table 1.
Summary of small-RNA sequencing data in three P. szechuanica libraries
| Type | Sb052 | Th053 | Control | |||
|---|---|---|---|---|---|---|
| Count | Percent | Count | Percent | Count | Percent | |
| total_reads | 17801357 | 19602945 | 17962927 | |||
| high quality | 17754456 | 100 % | 19544426 | 100 % | 17918326 | 100 % |
| 3’adapter_null | 3591 | 0.02 % | 5632 | 0.03 % | 2971 | 0.02 % |
| Insert_ null | 3869 | 0.02 % | 4910 | 0.03 % | 2943 | 0.02 % |
| 5’adapter_contaminants | 75547 | 0.43 % | 98975 | 0.51 % | 98892 | 0.55 % |
| smaller_than_18nt | 112591 | 0.63 % | 35223 | 0.18 % | 47711 | 0.27 % |
| Poly-A | 1373 | 0.01 % | 1284 | 0.01 % | 1334 | 0.01 % |
| clean reads | 17557485 | 98.89 % | 19398402 | 99.25 % | 17764475 | 99.14 % |
The small RNA (sRNA) reads were typically 19 to 25 nt in length (Fig. 1). Among these sequences, 21 nt sRNAs were the most abundant in the three libraries, accounting for 57.15 (Control), 52.55 (Sb052) and 48.34 % (Th053) of the total reads, followed by 24 nt sRNAs, which accounted for 17.13, 18.76 and 19.77 % of the total reads, respectively. These results were consistent with previous studies in Populus [24, 25, 27, 28]. However, it was also reported that the most abundant reads length was 24 nt, followed by 21 nt [31, 32], indicating that the miRNAs in Populus are primarily 21 and 24 nt in length. Furthermore, we observed that the number of 21 nt sequences in the control libraries was more abundant than that in the treatment libraries, and the number of 24 nt sequences in the treatment libraries was more abundant than that in the control libraries. The results suggest that the number of 21 nt miRNA sequences decreased and the number of 24 nt miRNA sequences increased in P. szechuanica after infection with foliar rust fungus.
Fig. 1.
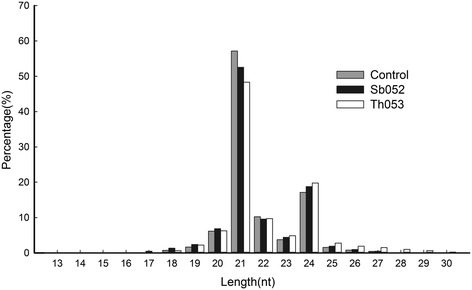
Length, distribution and frequency of sRNAs in the three P. szechuanica libraries
More than ten million total sRNA sequences and nearly three million unique sRNA sequences were identified (Table 2). Obviously, the sRNA sequences in the three libraries were sufficiently abundant to provide adequate data for further analysis. Unannotated sequences accounted for 30.99, 30.56 and 32.33 % of the total clean reads and 81.78, 80.95 and 81.21 % of the unique reads in Sb052, Th053 and control libraries, respectively. Thus, unknown sequences accounted for substantial proportions of the RNA categories in each library.
Table 2.
Distribution of sRNAs in different categories in three libraries of P. szechuanica
| Category | Sb052 | Th053 | Control | |||
|---|---|---|---|---|---|---|
| Unique sRNAs (%) | Total sRNAs (%) | Unique sRNAs (%) | Total sRNAs (%) | Unique sRNAs (%) | Total sRNAs (%) | |
| Total | 2905624(100) | 17557485(100) | 3287847(100) | 19398402(100) | 2797933(100) | 17764475(100) |
| exon | 136541 (4.70) | 576314(3.28) | 139463(4.24) | 553903(2.86) | 136964(4.89) | 674443(3.80) |
| intron | 109233(3.76) | 455077(2.59) | 111882(3.40) | 441890(2.31) | 109735(3.93) | 472656(2.66) |
| miRNA | 35186(1.21) | 7360137(41.92) | 38672(1.18) | 7664446(39.51) | 36008(1.29) | 7911943(44.54) |
| rRNAetca | 247466(8.52) | 3721683(21.2) | 335307(10.21) | 4799682(24.75) | 242107(8.65) | 2958184(16.65) |
| repeat | 854(0.03) | 3168(0.02) | 968(0.03) | 3076(0.02) | 874(0.03) | 3268(0.02) |
| unannb | 2376344(81.78) | 5441106(30.99) | 2661555(80.95) | 5929105(30.56) | 2272245(81.21) | 5743981(32.33) |
arRNAetc included rRNA, snRNA, snoRNA and tRNA
bunannotated reads
Known miRNAs in P. szechuanica
Known miRNAs in P. szechuanica infected with M. larici-populina were identified through homologous alignment analysis using the plant miRNA in miRBase 20.0. A total of 90 known miRNAs were obtained in the Sb052, Th053 and control libraries, belonging to 42 families from 313 genomic loci (Additional file 1: Table S1). The percentages of dominant miRNA families are shown in Fig. 2a, and most of these miRNAs were largely conserved in various plant species, except miR6421, which was only identified in P. trichocarpa. The expression levels of a few miRNA families, such as MIR156 and MIR166, were obviously high in three libraries. The most abundant miRNA family was MIR156, accounting for 58.37, 63.61 and 54.02 % of the total sequences in Sb052, Th053 and control libraries, respectively.
Fig. 2.
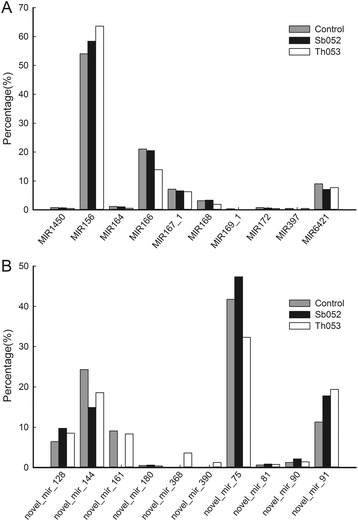
Abundance of highly expressed miRNAs in the three P. szechuanica libraries. a The percentages of 10 known miRNA families accounting for the total known reads. b The percentages of 10 novel miRNAs accounting for the total novel reads
miRNAs have a broad range of expression, varying from several to millions of reads. For example, miR157 generated 3,196,614 reads, whereas miR2275 generated only 11 reads in the control library. Most of the conserved miRNAs were identified from the three libraries, and certain miRNAs were abundant in some samples but scarce or even lacking in other samples. For example, the expression of miR4414 in Sb052 and Th053 libraries generated 82 and 118 reads, respectively; however, this miRNA was not present in the control library. Similarly, although the expression of miR2118 generated 7532 reads in the control library, this miRNA generated no reads in the other two libraries. Moreover, the number of reads for different members of same family varied widely. For example, miR473 and miR477 both belong to the miR477 family; however, 17,922 reads were obtained for miR473, whereas only 633 reads were obtained for miR477 in the control library.
A total of 70 miRNAs were common to the three libraries examined. Three miRNAs (miR5172, miR6446 and miR6465) were only identified in the Sb052 library, whereas four miRNAs (miR1863, miR476, miR6300 and miR7825) were specific to the Th053 library (Fig. 3a).
Fig. 3.
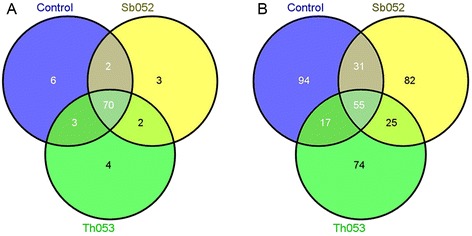
Venn diagrams of the miRNAs identified in the three libraries. a Known miRNAs. b Novel miRNAs
Novel miRNAs in P. szechuanica
The unique difference between miRNA and other small-molecule RNAs is that miRNA precursors form stem-loop structures as the basis for the prediction of new miRNA. In the present study, 378 novel miRNAs from 508 genome loci were identified using Mireap software (Beijing Genomics Institute (BGI), Shenzhen, China) (Additional file 2: Table S2). The precursor sequences and predicted secondary structures of 20 novel miRNAs are shown in Additional file 3: Figure S1. A total of 56 miRNAs were generated from multiple loci, among which novel_mir_16 had 13 loci. Marked differences in the abundance of the novel miRNAs were also observed. The percentages of highly expressed novel miRNAs are shown in Fig. 2b. The most abundant miRNA was novel_mir_75, with 59,661, 35,565 and 81,373 reads corresponding to 47.36, 32.32 and 41.77 % of the total reads in the Sb052, Th053 and control libraries, respectively, followed by novel_mir_144 and novel_mir_91. As shown in Fig. 3b, 55 novel miRNAs were common to the three libraries, whereas 82 miRNAs were unique to the Sb052 library and 74 miRNAs were only identified in the Th053 library.
Differential expression of miRNAs within inoculation treatments
The analysis of the differential expression of miRNAs in the inoculated and control libraries showed that 32 miRNAs (5 known and 27 novel miRNAs) were up-regulated and 40 miRNAs (15 known and 25 novel miRNAs) were down-regulated in P. szechuanica infected with Sb052, whereas 35 miRNAs (12 known and 23 novel miRNAs) were up-regulated and 53 miRNAs (15 known and 38 novel miRNAs) were down-regulated in P. szechuanica infected with Th053. The comparison of the miRNA expression levels in P. szechuanica inoculated with Sb052 and Th053 showed that 24 known (6 up-regulated and 18 down-regulated) and 52 novel miRNAs (33 up-regulated and 19 down-regulated) were differentially expressed between the two libraries. In brief, we identified 38 known and 92 novel differentially expressed miRNAs from the three libraries that might play important roles in response to pathogen stress (Additional file 4: Table S3). Among these miRNAs, miR2118, miR482, novel_mir_166 and novel_mir_198 were suppressed or down-regulated, while miR472, novel_mir_250 were up-regulated in both Sb052 and Th053 libraries compared to the control library. The expression level of miR1444, miR397 and novel_mir_191 were down-regulated in the Sb052 library, but their sequencing reads did not vary significantly in the Th053 library. In contrast, expression levels of novel_mir_248, novel_mir_290 and novel_mir_357 were up-regulated in the Sb052 library, but these miRNAs were not expressed in either the Th053 or the control library. The expression levels of miR164, novel_mir_11, novel_mir_211 and novel_mir_244 did not vary significantly in the Sb052 library, however, in the Th053 library, they were suppressed. In conclusion, the expressions levels of the same miRNAs were significantly different, and the expression levels of poplar miRNAs were more suppressed during rust infection.
Target prediction and functional analysis
The majority of plant miRNAs were completely complementary with their targets, which provided a convenient starting point for the prediction of target genes. A large number of targets were predicted for most miRNAs using the program developed by BGI (Shenzhen, China) and obeying the criteria and methods outlined previously [33, 34]. The results of KEGG pathway analysis revealed that target genes of these miRNAs were involved in various biological and biochemical processes in plant growth and development. The top ten preferential pathways of the target genes in the three P. szechuanica libraries were shown in Fig. 4. As for the known miRNAs, the pathway that accounted for the largest percentage of the total targets was metabolic pathways, followed by plant-pathogen interaction (Fig. 4a). However, regarding novel miRNAs, the pathway with the highest proportion of target genes was plant-pathogen interaction. Metabolic pathways took second place (Fig. 4b). Differentially expressed miRNAs and their target gene functional annotations were list in Additional file 4: Table S3. Among these miRNAs, twenty-seven miRNAs including 7 known miRNAs and 20 novel miRNAs in the leaves of P. szechuanica infected with M. larici-populina, were analysed and the results showed that the target genes were primarily associated with disease-resistance proteins, serine/threonine protein kinases, transcription factors, and related proteins (Table 3). The genes of disease resistance proteins RPS2, RPS5 and RPM1 were putative targeted by 3 known miRNAs (miR2118, miR472, miR482) and 8 novel miRNAs (novel_mir_11, novel_mir_166, novel_mir_211, novel_mir_244, novel_mir_248, novel_mir_250, novel_mir_357 and novel_mir_403). LRR receptor-like serine/threonine-protein kinase (FLS2) was predicted to be only targeted by several novel miRNAs, novel_mir_191, novel_mir_198, novel_mir_368, novel_mir_93. Two transcription factors involving in plant defense responses, transcription factor WRKY and MYC2, were found to be targeted by novel_mir_290 and miR530, respectively. In addition, polyphenol oxidase, L-ascorbate oxidase and peroxidase genes were also predicted to be targeted by miR1444, miR397 and novel_mir_191, respectively. Meanwhile, we also observed that some miRNAs have identical and unique targets; for example, the targets of the miR472, miR482, miR2118, novel_mir_11 and novel_mir_357 were all disease-resistance proteins, whereas some miRNAs, such as miR530, novel_mir_191, novel_mir_248, novel_mir_250 and novel_mir_270 have multiple target genes and functional annotations, including the genes of various enzymes, disease-resistance proteins and transcription factors.
Fig. 4.
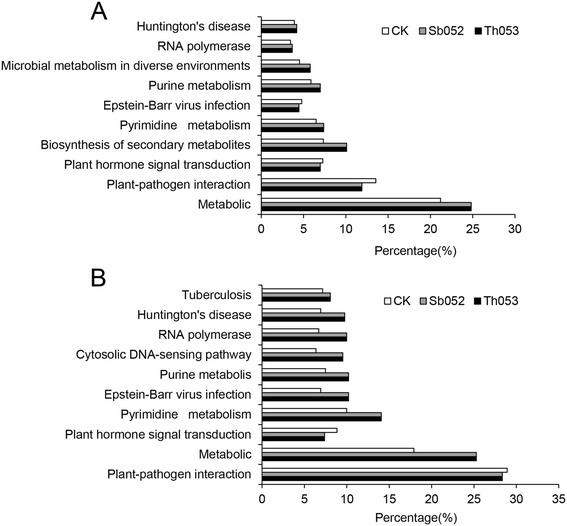
Top ten preferential pathways from KEGG pathway analysis of the target genes in the three P. szechuanica libraries. a Known miRNAs. b Novel miRNAs. X-axis represents the percentages of target genes involved in the ten pathways accounting for the total targets
Table 3.
Disease resistance-related miRNAs from P. szechuanica and target gene functional annotations
| miRNA name | Target accession | Functional annotation |
|---|---|---|
| miR1444 | Potri.011G047300.1,Potri.011G108200.1,Potri.011G108300.1,Potri.001G387900.1,Potri.001G388100.1,Potri.001G388300.1,Potri.001G388400.1,Potri.001G388800.1,Potri.001G388800.2,Potri.001G388900.1,Potri.T061900.1,Potri.T062100.1,Potri.T062200.1 | polyphenol oxidase |
| miR2118 | Potri.T003600.1,Potri.T003900.1,Potri.T004100.2,Potri.T045200.1,Potri.T004200.1,Potri.018G136300.1,Potri.T014600.1,Potri.T012900.1 | disease resistance protein RPS2 |
| disease resistance protein RPS5 | ||
| Potri.T004100.1,Potri.005G041100.2,Potri.005G041300.1,Potri.005G041100.1,Potri.005G042400.2,Potri.005G042400.1,Potri.005G042900.1,Potri.T016200.1,Potri.T016200.2 | disease resistance protein RPS2 | |
| Potri.T068700.1,Potri.011G011900.1,Potri.011G013800.1 | disease resistance protein RPM1 | |
| Potri.011G013100.1,Potri.011G015300.1 | disease resistance protein RPM1, | |
| disease resistance protein RPS2 | ||
| Potri.008G220200.1,Potri.008G220200.2 | disease resistance protein RPM1 | |
| disease resistance protein RPS5 | ||
| miR394 | Potri.018G095700.1,Potri.018G095700.2 | glycerol kinase |
| miR397 | Potri.010G183600.1,Potri.010G183600.2,Potri.009G034500.1,Potri.009G042500.1,Potri.009G102700.1,Potri.009G156600.1,Potri.009G156800.1,Potri.004G156400.2,Potri.004G156400.1,Potri.011G120200.1,Potri.011G120300.1,Potri.006G087100.1,Potri.006G094100.1,Potri.006G096900.1,Potri.006G097000.1,Potri.006G097100.1,Potri.016G106000.1,Potri.016G107500.1,Potri.016G112000.2,Potri.016G112000.1,Potri.016G112100.1,Potri.001G054600.1,Potri.001G184300.1,Potri.001G248700.1,Potri.001G401100.1,Potri.001G401300.1 | L-ascorbate oxidase |
| miR472 | Potri.T003600.1,Potri.T003900.1,Potri.T004100.2,Potri.T004200.1,Potri.T045200.1,Potri.T012900.1,Potri.018G136300.1,Potri.T014600.1 | disease resistance protein RPS2 |
| disease resistance protein RPS5 | ||
| Potri.T068700.1,Potri.011G013800.1 | disease resistance protein RPM1 | |
| Potri.011G013100.1,Potri.011G015300.1 | disease resistance protein RPM1 | |
| disease resistance protein RPS2 | ||
| Potri.T016200.1,Potri.T016200.2,Potri.T004100.1 | disease resistance protein RPS2 | |
| miR482 | Potri.017G015300.2,Potri.017G015300.1,Potri.017G015600.2,Potri.017G015600.4,Potri.017G015600.3,Potri.017G015600.1,Potri.004G196100.1,Potri.003G199100.1,Potri.003G199600.1,Potri.003G200800.1 | disease resistance protein RPM1 |
| Potri.001G427700.2,Potri.001G427700.1,Potri.001G432200.1 | disease resistance protein RPS2 | |
| Potri.001G432200.1 | disease resistance protein RPS5 | |
| miR530 | Potri.014G099700.1 | transcription factor MYC2 |
| novel_mir_11 | Potri.004G169800.1,Potri.004G169800.2,Potri.004G170300.1,Potri.003G017200.1,Potri.003G026900.1,Potri.018G136100.1,Potri.014G001900.1,Potri.014G002000.1,Potri.014G002200.1,Potri.014G002300.1,Potri.014G003200.2,Potri.014G003200.1,Potri.014G003400.1,Potri.014G003600.2,Potri.014G003600.1,Potri.014G005600.2,Potri.014G005600.1,Potri.014G007600.1,Potri.014G009300.1,Potri.014G009400.1,Potri.014G009400.2,Potri.014G009600.1,Potri.014G010600.1,Potri.014G010700.1,Potri.014G010900.1,Potri.014G012000.1 | disease resistance protein RPM1 |
| novel_mir_118 | Potri.006G047300.1,Potri.006G047300.2 | calcium-binding protein CML |
| novel_mir_166 | Potri.013G098000.1,Potri.013G097900.1 | disease resistance protein RPM1 |
| novel_mir_191 | Potri.018G035100.1 | LRR receptor-like serine/threonine-protein kinase FLS2 |
| Potri.019G103900.1,Potri.019G103900.2,Potri.019G103900.3 | serine/threonine-protein kinase PBS1 | |
| Potri.009G115200.1,Potri.009G115200.2 | brassinosteroid insensitive 1-associated receptor kinase 1 | |
| novel_mir_198 | Potri.005G031900.1 | LRR receptor-like serine/threonine-protein kinase FLS2 |
| novel_mir_244 | Potri.017G103500.1,Potri.017G103500.2, Potri.014G063500.1 | disease resistance protein RPM1 |
| Potri.014G064300.1 | disease resistance protein RPM1 | |
| disease resistance protein RPS2 | ||
| disease resistance protein RPS5 | ||
| novel_mir_248 | Potri.T049000.1 | disease resistance protein RPM1 |
| disease resistance protein RPS2 | ||
| novel_mir_206 | Potri.012G054700.1,Potri.012G054700.2 | chitin elicitor receptor kinase 1 |
| novel_mir_211 | Potri.017G127200.1,Potri.017G127800.1,Potri.017G128000.2,Potri.017G128000.1,Potri.017G133600.1,Potri.017G133700.1,Potri.017G136400.1,Potri.017G136700.1,Potri.017G136900.1,Potri.017G138100.1,Potri.018G080500.1,Potri.018G080700.1,Potri.T105500.1,Potri.001G033100.1,Potri.T096100.1 | disease resistance protein RPM1 |
| novel_mir_222 | Potri.013G108200.2 Potri.013G108200.1 | cyclic nucleotide gated channel, other eukaryote |
| novel_mir_25 | Potri.012G054700.1,Potri.012G054700.2 | chitin elicitor receptor kinase 1 |
| novel_mir_250 | Potri.019G018800.1,Potri.T068600.1,Potri.011G008600.1,Potri.011G011600.1,Potri.011G060600.1,Potri.011G008700.1,Potri.011G013400.1,Potri.011G015400.1,Potri.006G269900.1 | disease resistance protein RPM1 |
| Potri.T037600.1,Potri.T039900.1,Potri.T039100.1,Potri.T039300.1,Potri.006G270000.1 | disease resistance protein RPS2 | |
| disease resistance protein RPM1 | ||
| Potri.T067500.1 | disease resistance protein RPM1 | |
| disease resistance protein RPS5 | ||
| Potri.T112200.1,Potri.001G363400.1 | disease resistance protein RPM1 | |
| disease resistance protein RPS2 | ||
| novel_mir_270 | Potri.005G231100.2 | extracellular signal-regulated kinase 1/2 |
| novel_mir_290 | Potri.001G361600.1,Potri.001G361600.2, Potri.001G361600.3 | WRKY transcription factor 33 |
| novel_mir_357 | Potri.T003600.1,Potri.T044800.1,Potri.T045300.2,Potri.T045300.2,Potri.T045300.1,Potri.T045300.1,Potri.T024700.1,Potri.T024900.1,Potri.T025100.1,Potri.T025800.1,Potri.T025800.1,Potri.T026400.1,Potri.T026800.1,Potri.T026700.1,Potri.T026900.1,Potri.T027200.1,Potri.T027500.1,Potri.T027300.1,Potri.T027700.1,Potri.T028100.1,Potri.T028300.1,Potri.019G022800.1,Potri.019G020500.1,Potri.019G020200.2,Potri.019G020200.1,Potri.019G014500.1,Potri.018G137900.1,Potri.018G136500.1,Potri.018G135600.1,Potri.018G136700.1,Potri.T015900.2,Potri.T015900.1,Potri.T013900.1,Potri.T013800.1,Potri.T013300.3,Potri.T013300.2,Potri.T053000.1,Potri.001G406000.1,Potri.T012000.1,Potri.T013300.1,Potri.T014600.1 | disease resistance protein RPS2 |
| disease resistance protein RPS5 | ||
| Potri.T045000.1,Potri.T046000.1,Potri.T025300.1,Potri.T026200.1,Potri.T026600.1,Potri.011G127300.2,Potri.T028700.1,Potri.T028000.1,Potri.011G127300.1,Potri.T014900.1,Potri.019G002800.1,Potri.019G002800.2 | disease resistance protein RPS2 | |
| novel_mir_368 | Potri.011G104700.1,Potri.011G104900.1,Potri.T085700.2,Potri.T085700.3,Potri.T085700.1,Potri.T086100.3,Potri.T086100.1,Potri.T086100.2 | LRR receptor-like serine/threonine-protein kinase FLS2 |
| novel_mir_38 | Potri.007G128600.1 | calcium-binding protein CML |
| novel_mir_387 | Potri.004G025200.1,Potri.004G026100.1 | serine/threonine-protein kinase PBS1 |
| novel_mir_403 | Potri.T001600.1 | disease resistance protein RPM1 |
| novel_mir_93 | Potri.003G041600.1 | LRR receptor-like serine/threonine-protein kinase FLS2 |
QRT-PCR validation of P. szechuanica miRNAs
To confirm the temporal dynamic expression of miRNAs involved in pathogen stress, we further analysed 10 miRNAs in P. szechuanica infected with M. larici-populina at 0, 12, 24, 48, 96, and 144 hpi using qRT-PCR. The results showed that the miRNA expression levels dramatically changed during the pathogenesis of foliar rust infection (Fig. 5). When plants were infected with Sb052, the miRNA expression levels exhibited three patterns: Pattern A, including miR1444, miR398, novel_mir_25, and novel_mir161, gradually increased at the early stage, peaked at 24 or 48 hpi and then gradually declined; Pattern B, including miR482, novel_mir_180, novel_mir_357, and novel_mir_368, initially increased, then decreased, and subsequently increased again, peaking at 96 hpi; Pattern C, including miR394 and miR6462, initially decreased, then increased, peaking at 48 hpi, and subsequently declined again. When plants were infected with Th053, the miRNA expression levels were classified into four patterns. The first three patterns were identical to those observed with Sb052: Pattern A included miR394 and novel_mir_161; Pattern B included miR1444, miR482, novel_mir_180, and novel_mir_368, whose expression markedly declined, reaching the lowest point at 48 hpi; and Pattern C only included novel_mir_357. Pattern D, including miR398, miR6462 and novel_mir_25, suddenly increased, peaking at 12 or 96 hpi, followed by a dramatic decline. In the present study, the sequencing results showed that novel_mir_368 and novel_mir_161 were only observed in the Th053 library or in both the Th053 library and control, respectively; however, these miRNAs could be detected in the Sb052 library using qRT-PCR analysis. These different results suggest that deep sequencing was not sufficient for the detection of the expression of all miRNAs. Interestingly, we observed that the expression of 10 miRNAs, except miR394, novel_mir_161, and novel_mir_357, increased at 48 hpi in the Sb052 library and decreased at 48 hpi in the Th053 library compared with expression levels at 0 hpi, indicating that the expression levels of the same miRNAs differed in incompatible and compatible interaction libraries.
Fig. 5.
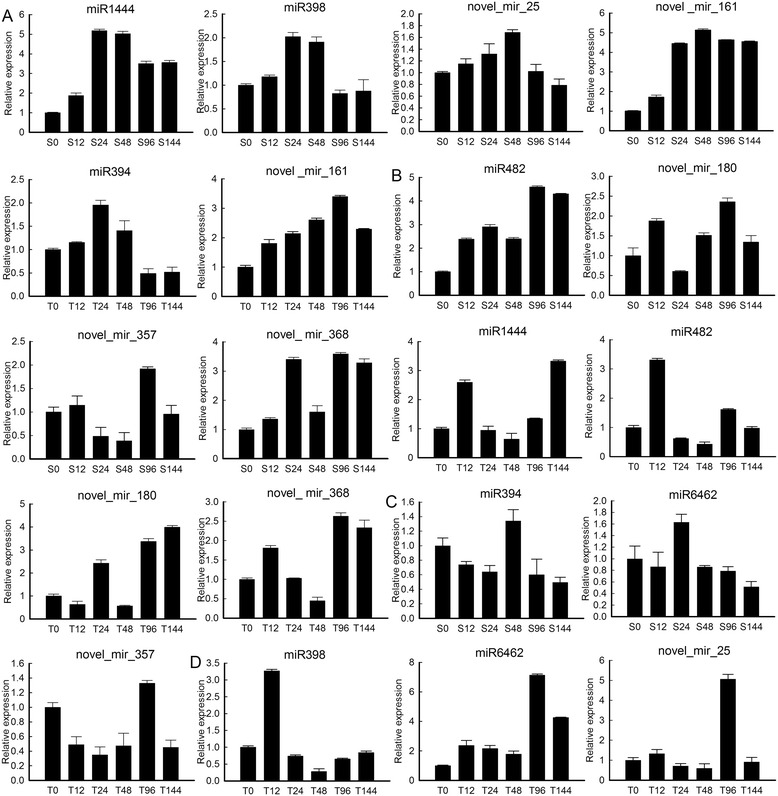
Temporal dynamic expression of miRNAs in P. szechuanica infected with M. larici-populina at 0, 12, 24, 48, 96 and 144 hpi. When plants were infected with Sb052(S), the validated miRNAs were classified into expression patterns a, b and c. When plants were infected with Th053 (T), the validated miRNAs were classified into expression patterns a, b, c and d. For normalization, 5.8S rRNA was selected as a reference gene, and the expression level at 0 hpi (S0 and T0) was set to 1. The statistical analyses and graphics were generated using the SigmaPlot 12.5 statistical package
Discussion
Identification of miRNAs in P. szechuanica
Several studies concerning miRNA expression in Populus subjected to various stresses have recently been conducted, and the number of miRNAs identified in Populus is increasing. A total of 405 mature Populus miRNA sequences have been registered in miRBase 20.0, among which 401 miRNAs have been identified in P. trichocarpa and 4 miRNAs have been identified in P. euphratica. However, there are few studies concerning the expression of miRNAs under pathogen stress in Populus. In the present study, we used high-throughput sequencing to construct libraries from the uninfected leaves of P. szechuanica and leaves infected with M. larici-populina avirulent isolate Sb052 and virulent isolate Th053. Sequencing obtained 90 known miRNAs belong to 42 families (Additional file 1: Table S1) and 378 novel miRNAs (Additional file 2: Table S2). The known miRNAs were matched with miRBase 20.0, and 43 miRNAs were identified in Populus, of which 41 miRNAs were P. trichocarpa miRNAs and 2 miRNAs were specific to P. euphratica. The remaining miRNAs were conserved in other plant species. Among the conserved miRNAs, MiR156 and MiR166 families were the most abundant, similar to their expression levels in P. euphratica [26], P. tomentosa [28] and P. beijingensis [31], whereas novel miRNAs showed low expression levels compared with the known miRNAs in P. szechuanica. A majority of known miRNAs were common to the three libraries, but most of novel miRNAs were only identified in a certain libraries (Fig. 3). These results showed that the unique miRNAs in P. szechuanica were abundant and their expression levels were altered by M. larici-populina infection.
Expression and targets function of miRNAs under pathogen stress
In the present study, 32 up-regulated and 40 down-regulated miRNAs were identified in P. szechuanica infected with Sb052, whereas 35 up-regulated and 53 down-regulated miRNAs were identified in plants infected with Th053. These findings revealed that more miRNAs were suppressed during rust infection. Among these miRNAs, miR1444, miR482 and miR2118 were significantly down-regulated upon rust fungus infection, consistent with the role of these miRNAs as negative regulators of disease-resistance proteins [23, 31, 35]. Whereas previously reported expression levels of miR167 [36], miR169 [31], miR172 [30], miR398 [31] were contrary to the results obtained in the present study. This differential expression might reflect different miRNA functions in different poplar-pathogen interactions or different inoculation tissues and durations, suggesting that miRNAs have complex regulatory functions in plants subjected to biotic stresses.
According to the functional analysis of the target genes by KEGG pathway database, the high-expressed known miRNAs were mostly responsible for controlling the basic metabolism pathways, while the low-expressed novel miRNAs were mainly involved in regulating the plant-pathogen interaction (Fig. 4). We can see that plant-pathogen interaction and metabolic pathways were significantly over-represented in poplar upon rust fungal inoculation.
miRNAs and their target genes have multiple functions in different plant species. MiR156 was highly conserved and involved in various stresses. It target genes encoded the nucleotide binding site-leucine rich repeat (NBS-LRR) disease resistance protein [37] and squamosa promoter-binding protein (SPB), known to regulate plant development and growth [22, 30, 38]. However, in our trial, miR156 had 43 targets, among which 6 targets were predicted that encoded ubiquitin C, which is involved in DNA repair, translational control and signal transmission. The differentially expressed miRNAs and the functions of their target genes were shown in Additional file 4: Table S3. The expression of miR168 was up-regulated in P. beijingensis [31] and P. trichocarpa [30] inoculated with canker disease pathogen as well as P. szechuanica inoculated with virulent isolate Th053. Four target genes of miR168 encoded argonaute (AGO) which is closely involved in the biosynthesis and function of miRNA. AGO1 mRNA is involved in a negative feedback regulation through the action of miR168 [31, 39]. Previous reports of the expression of miR167 [36] and miR172 [30] were contrary to the results obtained in the present study. miR167 predicted 7 targets encoding auxin response factor (ARF), which is involved in plant hormone signal transduction pathway [30, 36]. Auxin signal transduction is related to bacterial disease resistance in Arabidopsis [40], and fungal disease resistance in P. trichocarpa [30]. One target of miR172 encoded APETALA2, which is involved in floral development in many plant species under different stresses [30, 37, 38] and another target gene in loblolly pine infected with fusiform rust encoded an LRR protein kinase [37]. In addition, the target functions of several miRNAs were unknown and the targets of some miRNAs have not been predicted.
To uncover the functions of miRNAs in P. szechuanica under rust fungus-induced stress, the miRNA target genes involved in disease resistance were annotated using the KEGG pathway database (Table 3). miR1444 expression was down-regulated in the Sb052 library. The target gene of miR1444 encoded polyphenol oxidase, which is an important enzyme for resistance to biotic and abiotic stresses [23, 32]. The over-expression of polyphenol oxidase in tomato enhanced resistance to the pathogen Pseudomonas syringae [41]. Similarly, expressions of miR397 were obviously down-regulated in the incompatible interaction. The target gene L-ascorbate oxidase influenced tobacco growth and defense responses by modulating redox state of the apoplastic ascorbic acid [42]. The expression of miR530 was down-regulated in the Sb052 library but up-regulated in the Th053 library, and the target genes of miR530 encoded the transcription factor MYC2, a member of the plant-pathogen interaction pathway, and 1-phosphatidylinositol-4-phosphate 5-kinase, involved in the defence response of poplars to M. larici-populina [43]. The target genes of 11 miRNAs, including 3 known miRNAs and 8 novel miRNAs encoded disease resistance proteins, including RPM1, RPS2 and RPS5. These targets all belong to the largest class of disease resistance proteins, the NBS-LRR class, which confer resistance to bacterial, fungal, or viral pathogens [44–46]. The Arabidopsis thaliana RPM1, RPS2 and RPS5 genes confer resistance to the bacterial pathogen P. syringae [47–49]. In previous studies, the target genes of miR472 [32, 37], miR482 [31, 37, 50] and miR2118 [33, 51] were also found to encode disease-resistance proteins. These findings imply that these miRNAs are responsive to disease stress in different plants and play similar roles in resistance to pathogen infections.
The results of the qRT-PCR analysis demonstrated that most miRNAs in P. szechuanica infected with the avirulent isolate Sb052 and virulent isolate Th053 were up-regulated and down-regulated at 48 hpi, respectively. miR1444 is important in plants under biotic and abiotic stresses [23]. The expression of miR1444 at 24 and 48 hpi was lower than that at other times in compatible interactions; however, miR1444 expression reached high levels at the same times in incompatible interactions. In previous studies, many miRNAs were suppressed during fungus infection in compatible interactions, whereas most miRNAs did not vary with respect to incompatible interactions [38] or the number of down-regulated miRNAs was slightly less than that observed in compatible interactions [35]. Rinaldi et al. [43] elucidated that a great number of transcripts were significantly differentially regulated in incompatible interactions, but almost no transcriptional changes were detected in compatible interactions at 48 hpi. These findings provide us with evidence that 48hpi is a vital period for poplar-rust fungus interactions, and related genes are expressed differentially. According to these results, we speculate that miRNAs could possibly regulate the expression of certain genes and, through the changing expression of miRNAs, could play vital roles in regulating disease-resistance in Populus infected with rust. Furthermore, a significant difference exists between compatible and incompatible interactions at 48hpi. Therefore, the regulated functions of miRNAs in P. szechuanica infected with virulent and avirulent rust isolates at 48 hpi are complex, and require further investigation to understand the molecular mechanism of poplar-rust interactions.
Conclusions
In summary, a total of 90 known miRNAs and 378 novel miRNAs were identified from P. szechuanica infected with the M. larici-populina incompatible isolate Sb052, the M. larici-populina compatible isolate Th053 and uninfected controls through high-throughput sequencing. Comparative analysis revealed that the expression of miRNAs was significantly different after infection with different rust isolates, and more miRNAs were suppressed during rust infection. The targets of 27 miRNAs were primarily associated with disease resistance. The qRT-PCR analysis showed that miRNA expression exhibited different temporal dynamics in incompatible and compatible libraries. The regulatory mechanism of poplar miRNAs in response to rust fungus infection should be further studied.
Methods
Plant materials and rust fungus inoculation
The urediniospores of M. larici-populina isolates were multiplied on 1- to 2-year-old potted P. purdomii plants susceptible to all M. larici-populina isolates. Based on a previous study, the wild poplar P. szechuanica from the Qinling Mountains was used as the experimental host. One-year-old cuttings from P. szechuanica branches were potted and grown in the greenhouse for two months, and subsequently, the abaxial surfaces of the leaves were inoculated with a urediniospore suspension (1–2 mg/mL) containing isolates Sb052 (pathotype 1−-1) and Th053 (pathotype 3+-4) of M. larici-populina, forming incompatible and compatible interactions, respectively. In addition, the leaves were treated with running water as a control. The inoculation method has been previously described in Rinaldi et al. [43]. The P. szechuanica leaves from plants under the three treatments were separately collected at 0, 12, 24, 48, 96 and 144 hpi, immediately frozen in liquid nitrogen and stored at−80 °C until further analysis.
Total RNA extraction and sRNA sequencing
Total RNA was extracted from the frozen leaf tissues of inoculated and uninoculated P. szechuanica using TRNzol reagent (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Total RNA from each treatment at six time points was pooled with equal quantities to identify as many miRNAs as possible. The pooled RNA was used for sRNA library construction for the three treatments, and sRNA was sequenced using an Illumina-Solexa 1G Genetic Analyzer at the BGI (Shenzhen, China).
Analysis of sequencing data
Clean reads were obtained from raw reads after removing low-quality and contaminant reads. The clean reads were mapped to the P. trichocarpa genome [52] using SOAP software [53]. The non-coding RNAs, including rRNAs, scRNAs, snoRNAs, snRNAs, and tRNAs deposited in the NCBI GenBank database [54] and Rfam (11.0) database [55], were removed. Small RNAs corresponding to the exons and introns of mRNA and repeat sequences were also excluded from further analysis. The remaining sRNA sequences were aligned to the miRBase 20.0 database [56], with a maximum of two mismatches, to identify known miRNAs in P. szechuanica. The obtained sequences were used to predict hairpin structures using the BGI (Shenzhen, China) program. The remaining unannotated sRNAs were used to predict novel miRNAs using the prediction software Mireap [57].
Target prediction of miRNAs and functional analysis of the target genes
Target genes were predicted using the program developed by BGI (Shenzhen, China) and obeying the following rules referring to Allen et al. [33] and Schwab et al. [34]: no more than four mismatches between the sRNA and target gene, no more than two adjacent mismatches in the miRNA/target duplex, no adjacent mismatches at positions 2–12 of the miRNA/target duplex (5’ of miRNA), no mismatches at positions 10–11 of the miRNA/target duplex, no more than 2.5 mismatches at positions 1–12 of the miRNA/target duplex (5’ of miRNA), and the minimum free energy (MFE) of the miRNA/target duplex should be 75 % of the MFE of the miRNA bound to the perfect complement. Target genes were searched using Populus transcript information [58]. To better understand the roles of miRNAs in P. szechuanica under rust fungus-induced stress, the potential target functions were annotated using the KEGG pathway database [59].
Differential expression analysis of miRNAs
The miRNA reads in the three libraries were used to analyse differential expression and determine significant differences between the control and treatment libraries. The frequency of miRNAs in the three libraries was normalized to one million to reduce potential errors before calculating the fold-change, P-value and ratio. Normalized expression = (actual miRNA counts/total counts of clean reads)*1,000,000. Fold-change = log2 (miRNA normalized read counts in the treatment library/miRNA normalized read counts in the control library.)
The P-value was calculated as follows:
In this formula, N1 and N2 represent the total counts of clean reads in control and treatment library, respectively, and x and y represent the counts of clean reads of a given miRNA in control and treatment libraries, respectively.
A fold change greater than 1 or less than −1 and a P-value less than 0.01 suggested that the difference in the miRNA expression between two libraries was highly significant. When the fold-change was greater 1 or less than−1 and the P-value was between 0.01 and 0.05, the expression of the miRNA was significantly different between the two libraries.
The ratio of miRNA normalized read counts in treatment library/miRNA normalized read counts in control library was used to determine changes in the expression of an miRNA in the treatment samples compared with the control sample. When the ratio was more than 2, the miRNA was up-regulated, and when the ratio was less than 1/2, the miRNA was down-regulated.
Quantitative real-time PCR analysis of miRNA expression
To verify the expression levels of identified miRNAs, qRT-PCR was performed in accordance with Shi and Chiang [60]. Total RNA was extracted from Sb052- and Th053-infected P. szechuanica leaves at 0, 12, 24, 48, 96 and 144 hpi, and qRT-PCR was performed using the IQ5 Real-time Quantitative PCR Detection System (Bio-RadLaboratories, California, USA) with the SYBR® PrimeScript™ miRNA RT-PCR Kit (TaKaRa, Dalian, China) according to the manufacturers’ instructions. The reactions were performed in a total volume of 25 μL containing 2.0 μL of diluted cDNA, 1 μL of each primer, and 12.5 μL of SYBR Green premix Ex Taq II with the following reaction conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 20 s. A melting curve analysis was generated to verify the specificity of the PCR amplification. Each sample was processed in triplicate. Ten miRNAs, including five known and five novel miRNAs, were tested. For normalization, the 5.8S rRNA was used as the reference gene [22] and the expression level at 0 hpi was set to 1. All validated miRNAs and primer sequences are listed in Additional file 5: Table S4.
Availability of supporting data
The supporting data are included within the article and additional files.
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (Project No. 30872027).
Abbreviations
- miRNAs
microRNAs
- qRT-PCR
quantitative real-time PCR
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- hpi
Hour post-inoculation
- nt
Nucleotides
- NBS-LRR
Nucleotide binding site-leucine rich repeat
- AGO
Argonaute
Additional files
Known miRNAs identified in the three P. szechuanica libraries. (XLSX 77 kb)
Novel miRNAs identified in the three P. szechuanica libraries. (XLSX 110 kb)
Precursor sequences and the predicted secondary structures of the 20 novel miRNAs from P. szechuanica (“(”represent base matches, “.” represent base mismatches). (PDF 387 kb)
Differentially expressed miRNAs from P. szechuanica and target gene functional annotations. (XLSX 31 kb)
miRNAs and primer sequences for qRT-PCR. (XLSX 10 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZMC and MC conceived and designed experiments. MC conducted experiments, analysed the data and drafted the manuscript. ZMC revised the manuscript and approved the final version of the manuscript for publication. Both authors have read and approved the final manuscript.
Contributor Information
Min Chen, Email: chenmin.11070@163.com.
Zhimin Cao, Email: zmcao@nwsuaf.edu.cn.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate starvation response in Arabidopsis. Curr Biol. 2005;15:2038–43. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–99. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Naya L, Paul S, Valdés-López O, Mendoza-Soto AB, Nova-Franco B, Sosa-Valencia G, et al. Regulation of copper homeostasis and biotic interactions by MicroRNA 398b in Common Bean. PLoS ONE. 2014;9:e84416. doi: 10.1371/journal.pone.0084416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He H, He L, Gu M. Role of microRNAs in aluminum stress in plants. Plant Cell Rep. 2014;33:831–6. doi: 10.1007/s00299-014-1565-z. [DOI] [PubMed] [Google Scholar]
- 6.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–19. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llave C. MicroRNAs: more than a role in plant development. Mol Plant Pathol. 2004;5:361–6. doi: 10.1111/j.1364-3703.2004.00227.x. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 9.Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol. 2010;48:225–46. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta OP, Sharm P, Gupta RK, Sharma I. Current status on role of miRNAs during plant-fungus interaction. Physiol Mol Plant Pathol. 2014;85:1–7. doi: 10.1016/j.pmpp.2013.10.002. [DOI] [Google Scholar]
- 11.Guo N, Ye W, Wu X, Shen D, Wang Y, Xing H, Dou D. Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome. 2011;54:954–8. doi: 10.1139/g11-050. [DOI] [PubMed] [Google Scholar]
- 12.Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.) BMC Plant Biol. 2010;10:123. doi: 10.1186/1471-2229-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma C, Lu Y, Bai S, Zhang W, Duan X, Meng D, Wang Z, Wang A, Zhou Z, Li T. Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (Polden Delicious) Mol Plant. 2014;7:218–30. doi: 10.1093/mp/sst101. [DOI] [PubMed] [Google Scholar]
- 14.Viswanathan C, Anburaj J, Prabu G. Identification and validation of sugarcane streak mosaic virus encoded microRNAs and their targets in sugarcane. Plant Cell Rep. 2014;33:265–76. doi: 10.1007/s00299-013-1527-x. [DOI] [PubMed] [Google Scholar]
- 15.Yin X, Wang J, Cheng H, Wang X, Yu D. Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta. 2013;237:1213–25. doi: 10.1007/s00425-012-1835-3. [DOI] [PubMed] [Google Scholar]
- 16.Podda A, Simili M, Del Carratore R, Mouhaya W, Morillonc R, Maserti BE. Expression profiling of two stress-inducible genes encoding for miraculin-like proteins in citrus plants under insect infestation or salinity stress. J Plant Physiol. 2014;171:45–54. doi: 10.1016/j.jplph.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Tuskan GA, DiFazio SP, Teichmann T. Populus genomics is getting popular: the impact of the populus genome project on tree research. Plant Biol. 2004;6:2–4. doi: 10.1055/s-2003-44715. [DOI] [PubMed] [Google Scholar]
- 18.Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 19.Barakat A, Wall PK, DiLoreto S, Carlson JE. Conservation and divergence of microRNAs in Populus. BMC Genomics. 2007;8:481. doi: 10.1186/1471-2164-8-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klevebring D, Street NR, Fahlgren N. Genome-wide profiling of Populus small RNAs. BMC Genomics. 2009;10:620. doi: 10.1186/1471-2164-10-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puzey JR, Karger A, Axtell M, Kramer EM. Deep annotation of Populus trichocarpa microRNAs from diverse tissue sets. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0033034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S, Sun YH, Shi R, Clark C, Li L, Chang VL. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu S, Sun YH, Chiang VL. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131–51. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Ren Y, Zhang Y, Xu J, Sun F, Zhang Z, Wang Y. Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene. 2012;504:160–5. doi: 10.1016/j.gene.2012.05.034. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Zhang Y, Ren Y, Xu J, Zhang Z, Wang Y. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem Biophys Res Commun. 2012;417:892–6. doi: 10.1016/j.bbrc.2011.12.070. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Qin Y, Duan H, Yin W, Xia X. Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot. 2011;62:3765–79. doi: 10.1093/jxb/err051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Duan H, Li J, Deng X, Yin W. Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol Biol. 2013;81:525–39. doi: 10.1007/s11103-013-0010-y. [DOI] [PubMed] [Google Scholar]
- 28.Ren Y, Chen L, Zhang Y, Kang X, Zhang Z, Wang Y. Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct Integr Genomics. 2012;12:327–39. doi: 10.1007/s10142-012-0271-6. [DOI] [PubMed] [Google Scholar]
- 29.Qin Y, Duan Y, Xia X, Yin W. Expression profiles of precursor and mature microRNAs under dehydration and high salinity shock in Populus euphratica. Plant Cell Rep. 2011;30:1893–907. doi: 10.1007/s00299-011-1096-9. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, Jiang X, Zhang B, Su X. Involvement of microRNA-mediated gene expression regulation in the pathological development of stem canker disease in Populus trichocarpa. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0044968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Ren Y, Zhang Y, Xu J, Zhang Z, Wang Y. Genome-wide profiling of novel and conserved Populus microRNAs involved in pathogen stress response by deep sequencing. Planta. 2012;235:873–83. doi: 10.1007/s00425-011-1548-z. [DOI] [PubMed] [Google Scholar]
- 32.Shuai P, Liang D, Zhang Z, Yin W. Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genomics. 2013;14:233. doi: 10.1186/1471-2164-14-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen E, Xie Z, Gustafson A, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–21. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D, et al. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–27. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Yin Z, Li Y, Han X, Shen F. Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahlia inoculated cotton roots. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0035765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta OP, Permar V, Koundal V, Singh UD, Praveen S. MicroRNA regulated defense response in Triticum aestivum L. during Puccinia graminis f.sp. tritici infection. Mol Biol Rep. 2012;39:817–24. doi: 10.1007/s11033-011-0803-5. [DOI] [PubMed] [Google Scholar]
- 37.Lu S, Sun YH, Amerson H, Chiang VL. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 2007;51:1077–98. doi: 10.1111/j.1365-313X.2007.03208.x. [DOI] [PubMed] [Google Scholar]
- 38.Kulcheski FR, de Oliveira LFV, Molina LG. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics. 2011;12:307. doi: 10.1186/1471-2164-12-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE 1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–97. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–9. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Steffens JC. Over-expression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta. 2002;215:239–47. doi: 10.1007/s00425-002-0750-4. [DOI] [PubMed] [Google Scholar]
- 42.Pignocchi C, Kiddle G, Hernández I, Foster SJ, Asensi A, Taybi T, et al. Ascorbate Oxidase-dependent changes in the redox state of the apoplast modulate gene transcript accumulation leading to modified hormone signaling and orchestration of defense processes in Tobacco. Plant Physiol. 2006;141:423–35. doi: 10.1104/pp.106.078469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rinaldi C, Kohler A, Frey P, Duchaussoy F, Ningre N, Couloux A, et al. Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiol. 2007;144:347–66. doi: 10.1104/pp.106.094987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Liu X, Dai L, Wang G. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genomics. 2007;34:765–76. doi: 10.1016/S1673-8527(07)60087-3. [DOI] [PubMed] [Google Scholar]
- 45.Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 46.Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JDG. Molecular genetics of plant disease resistance. Science. 1995;268:661–7. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 47.Tornero P, Chao RA, Luthin WN, Goff SA, Dangl JL. Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell. 2007;14:435–50. doi: 10.1105/tpc.010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawawicz BJ. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell. 1993;5:865–75. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren R, Henk A, Mowery P, Holub E, Innes RW. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell. 1998;10:1439–52. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shivaprasad PV, Chen HM, Patel K, Bond DM, Sanros BA, Baulcombe DC. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–74. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jagadeeswaran G, Zheng Y, Li Y, Shukla LI, Matts J, Hoyt P, et al. Cloning and characterization of small RNAs from Medicago truncatulareveals four novel legume-specific microRNA families. New Phytol. 2009;184:85–98. doi: 10.1111/j.1469-8137.2009.02915.x. [DOI] [PubMed] [Google Scholar]
- 52.The Populus trichocarpa genome. ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Ptrichocarpa/assembly/Ptrichocarpa_210.fa.gz (2012). Accessed 7 August 2013.
- 53.Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 54.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2014;42:D32–7. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–32. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q, Jiang W. Mireap: MicroRNA discovery by deep sequencing. 2008. http://sourceforge.net/projects/mireap/. Accessed 7 August 2013.
- 58.The Populus trichocarpa transcript. ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Ptrichocarpa/annotation/Ptrichocarpa_210_transcript.fa.gz (2012). Accessed 7 August 2013.
- 59.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. BioTechniques. 2005;39:519–25. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]


