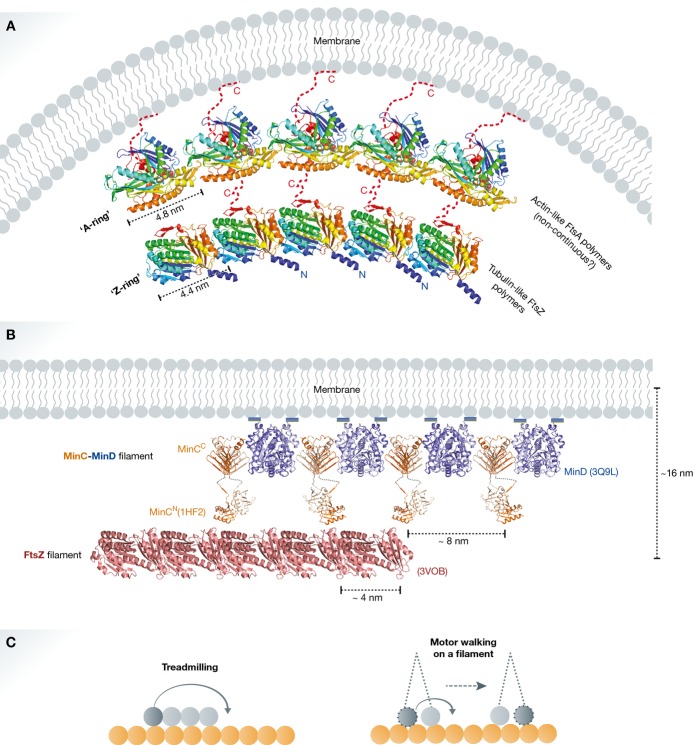
Figure 3. Two-protein collaborative filaments.
(A) FtsA is a bacterial actin homologue that recruits the nascent FtsZ ring to the cytoplasmic membrane. This is an example of a two-protein collaborative filament system on a matrix. There is a mismatch of repeat distances between FtsA and FtsZ polymers (∼5 nm and ∼4 nm, respectively). The mismatch causes filament curvature upon collaborative assembly (modified from Szwedziak et al, 2012). (B) MinCD cell division inhibitors form alternating copolymeric filaments on the membrane and interact with FtsZ filaments preventing polar cell division. This is another example of a two-protein collaborative filament system on a matrix. Here, the repeat distance of MinCD filaments (∼8 nm) is in multiples of the FtsZ filament repeat distance (∼4 nm). Hence, very little or no curvature is generated. This system shows cooperativity allowing MinC to attack FtsZ filaments over FtsZ monomers; thus, substoichiometric amounts of MinC can regulate Z-ring assembly (reproduced with permission from Ghosal et al, 2014). (C) Comparison between a two-filament collaborative filament system, where one cytomotive filament treadmills on another relatively stable filament (i) and a molecular motor walking on a filament (ii). If the movement of the lagging motor head to the front in the direction of the movement is assumed as treadmilling of a very short filament, consisting of only two subunits, then both systems have similar working principles. This postulation does not account for motor-to-motor communication, filament-induced nucleotide hydrolysis and the torque generation.