Figure 2.
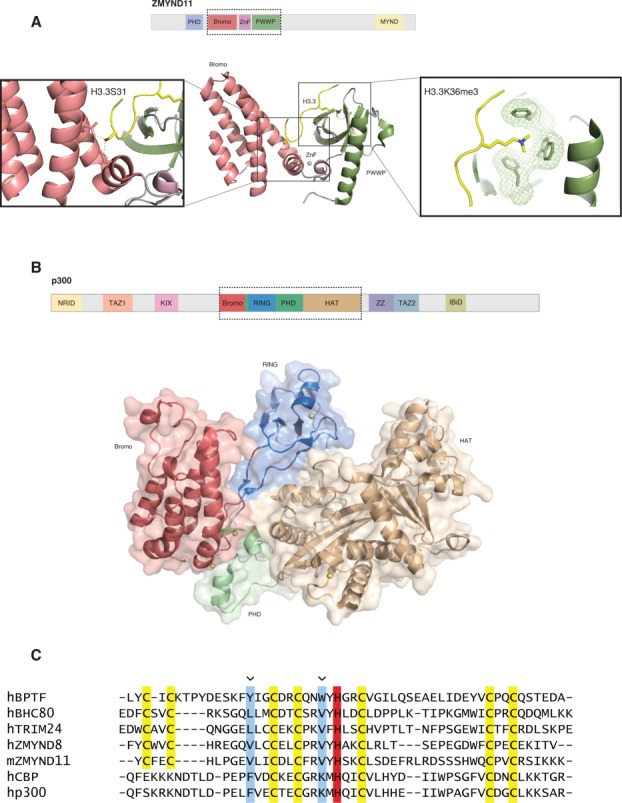
Structural studies reveal hidden zinc fingers in between tandem modules. (A) Top: Domain architecture of full-length ZMYND11. Bottom: Shown are the tandem bromodomain (red), zinc finger (purple), and PWWP (green) domains of ZMYND11. The histone H3 tail is shown in yellow. Visible in the left panel is the recognition of histone H3.3 specific residue S31 (H3.3S31). Recognition of this residue imparts the H3.3 specific binding of ZMYND11. The panel on the right illustrates binding of H3.3K36me3 by the “aromatic cage” of the PWWP domain. The “aromatic cage” is visible as mesh-covered sticks and H3.3K36me3 is visible as yellow sticks. (B) Top: Domain architecture of full-length p300. Bottom: Shown is the tandem bromodomain (red), RING (blue), PHD (green), and HAT (brown) module of P300. The RING domain inserts into the PHD domain. The PHD domain regulates catalytic activity of the adjacent HAT domain. (C) Sequence alignment shows that the PHD fingers of ZMYND8 and ZMYND11 have similar features to the H3K4me0 binding TRIM24 and BHC80 rather than the H3K4me3 binding BPTF. CBP and p300 lack the determinants for canonical H3K4 recognition. The zinc chelating cysteines are highlighted in yellow, the zinc chelating histidine is highlighted in red, the binding pocket residues are highlighted in blue and indicated with an arrow.
