Table II.
Residue-Residue Contacts Established Between the Peptides and eIF4E in the Computational Models
| Peptide's residue | Position | eIF4E residue | Peptide's residue | Position | eIF4E residue | |
|---|---|---|---|---|---|---|
| 4E-BPs and eIF4G peptides | CYFIP1 peptide | |||||
| MD model | BEDAM model | |||||
| Y | Y1 | His37, Pro38, Leu39, Gly139 | L | L1 | Ile138, Gly139 | |
| D/S/G | X2 | D | X2 | |||
| R/P | X3 | Glu132, Leu135, Arg186 | K | K3 | Asp143, Asp144 | Glu132, Glu140 |
| K/E/T | X4 | R | R4 | |||
| F | X5 | His37 | L | L5 | Val69, Trp73, Ile138 | |
| L | L6 | Val69, Trp73, Leu135, Ile138, Gly139 | R | R6 | Glu140, Ser141, Asp143 | Glu132, Leu135 |
| L/M |
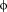 7 7 |
Trp73, Leu135 | S | S7 | ||
| D/E/G/Q | X8 | E | E8 | |||
| F/C/R | X9 | Val69, Glu70, Trp73 | C | C9 | Trp73, Leu135 | |
| K/R/Q | X10 | Trp73, Tyr76, Ile78, Glu128 | K | K10 | ||
| N/D/F | X11 | N | N11 | |||
aInteractions conserved among MD and X-ray or NMR structures are highlighted in bold.
Contacts occurring in more than 50% of the trajectory snapshots are listed. A contact is defined as an interatomic distance smaller than 4 Å.
