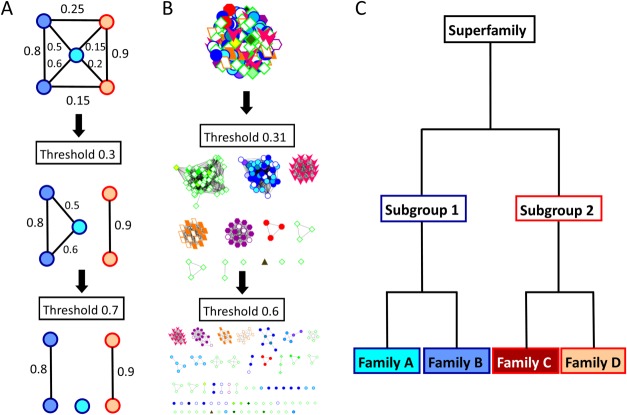
Figure 1.
Conceptual illustration of network-based clustering, which, ideally, would produce a functional hierarchy matching the SFLD. A. A similarity network is composed of nodes (proteins) connected to one another with edges (pairwise similarity scores). As the edge threshold is increased, all scores below that threshold are removed, producing distinct clusters. B. An actual similarity network hierarchy for the enolase superfamily. Each protein structure is represented by a node; pairwise ASP scores are represented by edges. This network is clustered into groups roughly mimicking subgroup annotations (border color and node shape) at a threshold of 0.31, but begins to break into specific families (fill color) and smaller groups at a threshold of 0.6. C. The SFLD defines a functional hierarchy, with superfamilies defined as sets of proteins which share a mechanistic step and the most detailed level, families, defined as sets of proteins which share entire mechanisms. Subgroups are illustrated with border color and families are illustrated with fill color.