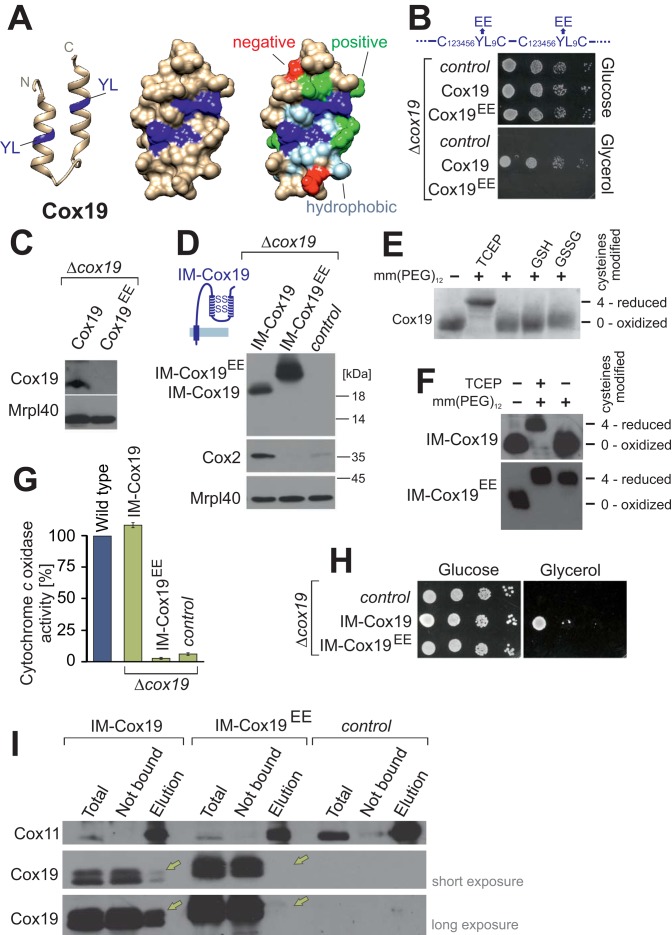
FIGURE 5:
Conserved tyrosine-leucine residues are essential for Cox19 activity. (A) Structure of the hydrophobic cavity formed by the twin Cx6YLxC motif of Cox19. The structure was modeled as in Figure 1A, but only the helix-loop-helix region is shown. YL dipeptides, dark blue; on rightmost structure, other hydrophobic residues, light blue; negative residues, red; and positive residues, green. (B) Cells from a Δcox19 strain were transformed with an empty plasmid (control) or plasmids overexpressing Cox19 or Cox19EE. Growth was analyzed on SD (glucose) or SG (glycerol) plates. (C) Western blot analysis of mitochondria (100 μg) isolated from Cox19- and Cox19EE-expressing cells. (D) Fusion proteins of an IMS-targeting sequence with Cox19 and Cox19EE were overexpressed in Δcox19 cells. Mitochondria were isolated and analyzed by Western blotting. (E) Cox19 was incubated with 5 mM TCEP, GSH, or GSSG, reisolated by TCA precipitation, and treated with the alkylating agent mm(PEG)12. Modification with mm(PEG)12 leads to a slower electrophoretic mobility, which is indicative of the presence of reduced thiols. Cox19 was fully oxidized unless treated with the strong reductant TCEP. (F) Mitochondrial proteins of the indicated strains were TCA precipitated and treated with TCEP and/or mm(PEG)12. (G) Activities of cytochrome c oxidase were analyzed in mitochondria isolated from the indicated strains. (H) Drop dilution experiment of the strains analyzed in D. (I) Cox11 was purified by immunoprecipitation with Cox11-specific antibodies from extracts of mitochondria isolated from the indicated strains. Copurified Cox19 was detected by Western blotting and is indicated by yellow arrows. Note that no Cox11 can be detected in the pull downs with IM-Cox19EE.