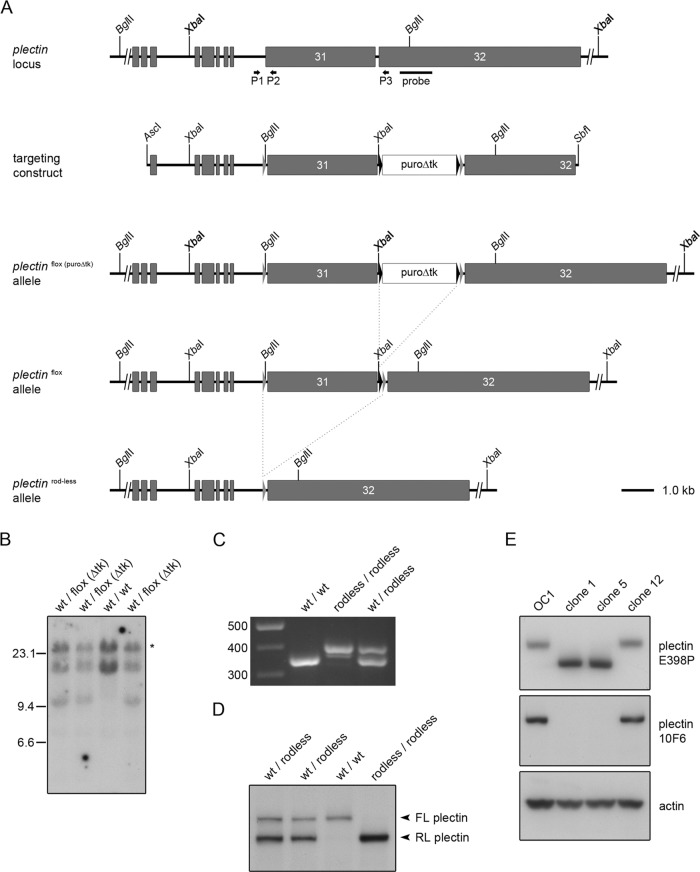
FIGURE 1:
Targeting strategy and molecular analysis of recombinant embryonic stem cells and rodless plectin mice. (A) Partial plectin gene structure, targeting construct, and different plectin mutant alleles. Gray boxes represent coding exons; gray and black triangles mark loxP and frt sites, respectively. Shown are the locations of the outermost 5′ and 3′ restriction sites used to generate the targeting construct and of the BglII and XbaI cleavage sites. The positions of the hybridization probe used for Southern blotting and the primers (arrows) used for the analysis of the different mutant alleles by PCR are indicated below the wild-type plectin allele. Dotted lines indicate the FLPe- and Cre-specific recombination events. (B) Southern blot analysis of four independently targeted ES cell clones. ES cell DNA was digested with XbaI (bold), subjected to agarose gel electrophoresis, and transferred to nitrocellulose. The 14.6 and 11.3-kb fragments corresponding to wild-type and floxed (puroΔtk) alleles, respectively, were detected by hybridization with a radiolabeled plectin genomic probe. The asterisk indicates the product of a partially digested wild-type allele. (C) PCR analysis of genomic DNA from wild-type, rodless plectin, and heterozygous mice using primers P1–P3. (D) Western blot analysis for the presence of full-length (FL) and rodless (RL) plectin in skeletal muscle tissue lysates from wild-type, rodless plectin, and heterozygous mice. (E) Western blot analysis of cell lysates from keratinocyte clones expressing full-length (OC1, clone 12) or rodless (clones 1 and 5) plectin. Different plectin antibodies recognizing epitopes outside (E398P) or within (10F6) the rod domain were used. Actin levels served as a loading control.