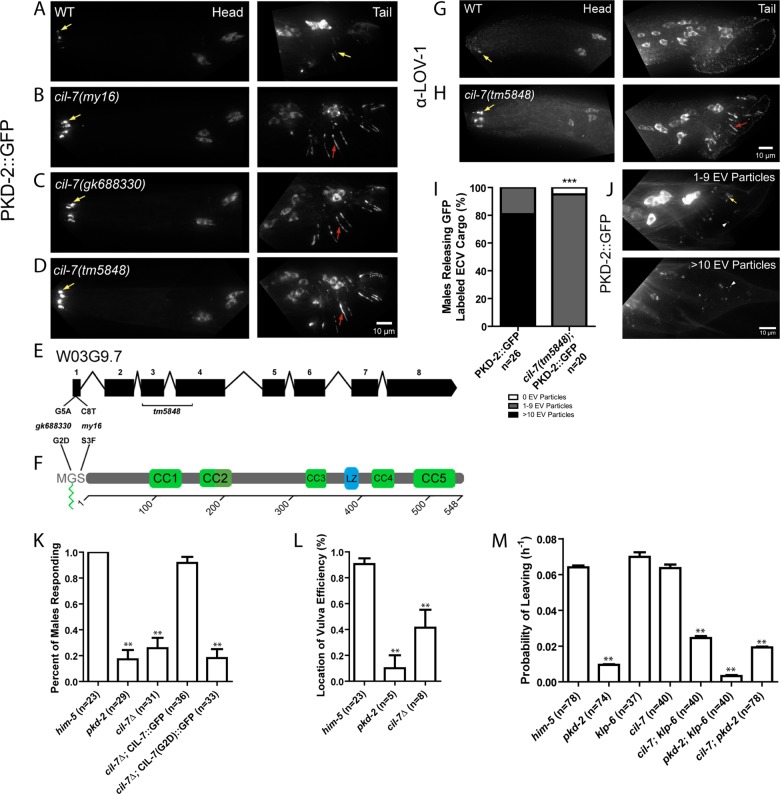
FIGURE 1:
cil-7 is required for the localization of the PCs and for male mating behaviors. (A) PKD-2::GFP localized to the cilia and cell bodies of the CEM, RnB, and HOB neurons of males. (B–D) In cil-7(my16), cil-7(gk688330) and cil-7(tm5848) males, head CEM neurons and tail HOB and RnB neurons accumulated PKD-2::GFP along the dendrites and cilia. (E) cil-7 genomic structure. my16 (C8 → T) and gk688330 (G5 → A) are missense SNPs. tm5848 is an out-of-frame deletion. (F) cil-7 encodes a predicted protein with an N-terminal myristoylation motif, five coiled-coil domains, and a leucine zipper. (G) The anti–LOV-1 antibody showed that endogenous LOV-1 localized to the cilia and cell bodies of the CEM, HOB, and RnB neurons. (H) In cil-7(tm5848) males, endogenous LOV-1 accumulated at the base and along the cilia of the CEM neurons. Excess LOV-1 appeared along the cilia and the dendrites of the RnB neurons. (I) In the cuticle of L4 molting wild-type males, >10 PKD-2::GFP–containing EV particles were observed in 80% of animals scored. In cil-7(tm5848) L4 molting males, much fewer PKD-2::GFP–containing EVs were observed. (J) Representative images of PKD-2::GFP EV particle ranges (1–9 or >10) observed to be trapped in the molted tail cuticle of late L4 males. (K) cil-7(tm5848) males did not respond to hermaphrodite contact. The translational reporter Pcil-7::CIL-7::GFP rescued the response defect of cil-7(tm5848) males. By contrast, the myristoylation-defective reporter Pcil-7::CIL-7(G2D)::GFP failed to rescue response defects of cil-7(tm5848) males. (L) cil-7(tm5848) males were location-of-vulva (Lov) defective. (M) cil-7(tm5848) males were not-leaving-assay defective (nonLas). The cil-7(tm5848); klp-6(my8) double mutant was Las, unlike the cil-7(tm5848) or klp-6(my8) single mutants. In I, ***p < 0.0001 by the Mann–Whitney test. In K, data were analyzed with Fisher's exact test between all groups, followed by Holm–Bonferroni multiple comparison adjustment with a α of 0.01 (**). In L, data were analyzed with the pairwise Mann–Whitney U-test between all groups, followed by Holm–Bonferroni multiple comparison adjustment with a total α of 0.01 (**). In M, PL values were compared by Holm–Bonferroni multiple comparison adjustment with a total α of 0.01 (**). Yellow arrow, cilia; red arrow, dendrite; white arrowhead, EV particle.