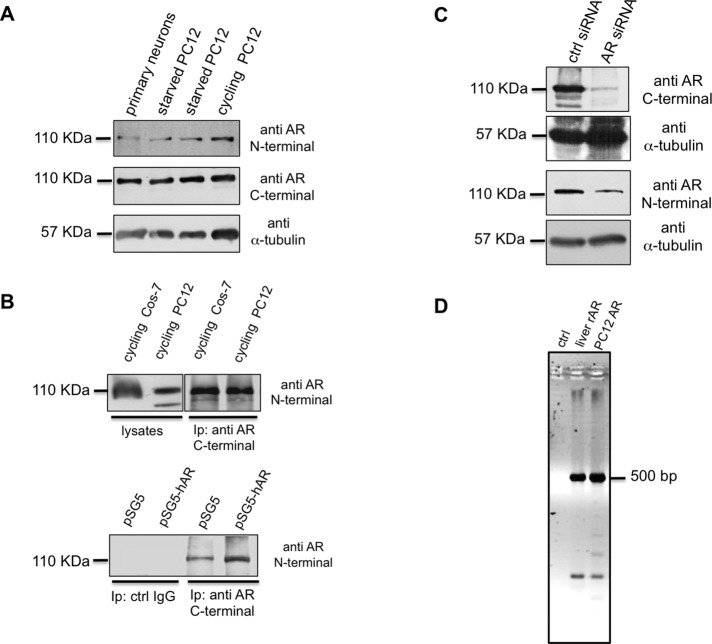
FIGURE 2:
Characterization of endogenous AR in PC12 cells. (A) Primary neurons and starved and cycling PC12 cell lysates were prepared as described in Materials and Methods and resolved on SDS–PAGE using 10% acrylamide. Proteins were transferred to nitrocellulose filter and probed with the antibodies against N- or C-terminal domains of AR. The filter was reprobed using anti–α-tubulin antibody as a loading control. (B) Top, lysate proteins from cycling Cos-7 ectopically expressing hAR or PC12 cells were used for IP experiments using an antibody raised against the AR C-terminal domain. Bottom, cycling PC12 cells were transfected with pSG5 or pSG5-hAR plasmid as described in Materials and Methods. Lysate proteins (2 mg/ml) were used for IP experiments using an antibody raised against the AR C-terminal domain. Proteins from cell lysates and immune complexes were resolved on SDS–PAGE using 8% acrylamide, transferred to nitrocellulose filter, and probed with the antibody against the N-terminal domain of AR. (C) Cycling PC12 cells were transfected with control (ctrl) or AR siRNA, as described in Materials and Methods. Cells were made quiescent, and lysate proteins were resolved on SDS–PAGE, using 10% acrylamide. Proteins were transferred to nitrocellulose filter and probed with the antibodies against C- or N-terminal domains of AR. The filters were reprobed, using anti–α-tubulin antibody, as a loading control. (D) PCR products using the primers set indicated in Materials and Methods were amplified from rat liver genomic DNA (liver rAR), PC12 genomic DNA (PC12 AR), or water (ctrl) and separated by electrophoresis in 2% agarose in TBE. A PCR product of ∼500 base pairs corresponding to the size expected from a wild-type sequence (see also data in Supplemental Figure S3) was amplified from both genomic DNA samples.