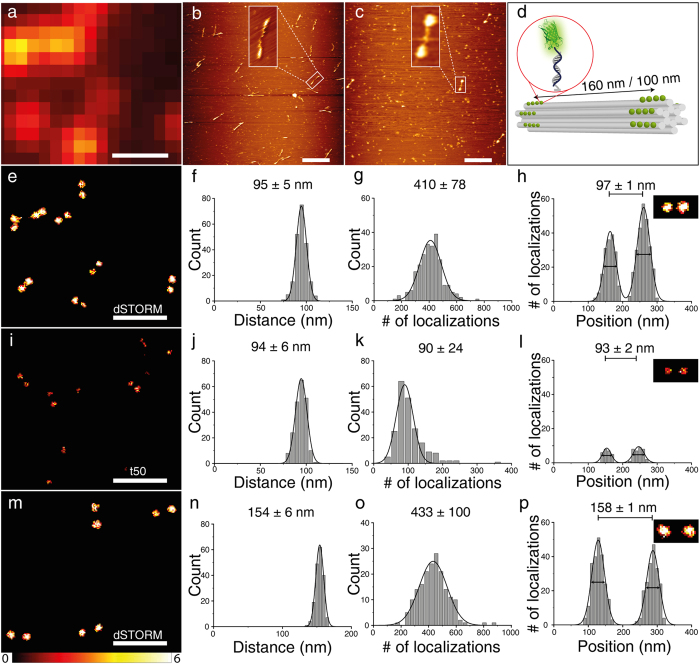
Figure 3.
(a) TIRF-image of 12 helix bundle DNA origamis with eYFP-DNA conjugates with a designed distance of 100 nm, scale bar 500 nm which corresponds to the super-resolved image in 3e. (b) AFM image of 12HB DNA origamis (160 nm). (c) AFM image of 12HB DNA origamis with eYFP-DNA conjugates (160 nm). Background spots are due to BSA used to increase filtering yields, scale bar 500 nm. (d) Sketch of 12HB DNA origamis with designed distances of 100 nm or 160 nm between two marks whereas each mark is created by 16–19 single stranded protrusions. Super-resolution imaging of eYFP labeled 12HBs (100 nm and 160 nm) was obtained by successive localization of blinking molecules (e–p)31. (e,i,m) Super-resolution images. (f,j,n) Histograms of intermark distances. (g,k,o) Overall number of localizations per 12HB DNA origami. (h,l,p) Cross sections of pixels indicated in inset in dSTORM-buffer (FWHM 39/42 nm in (h), 42/42 nm in (p)) and in t50-buffer (FWHM 31/38 nm in (l)), respectively. More than 200 structures were analyzed for each distance and buffer condition. Errors in (f–h,j–l,n–p) are standard deviation of Gaussian fits.