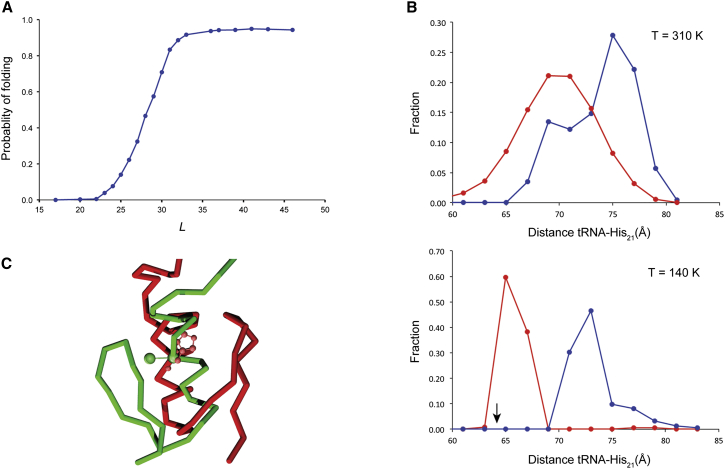
Figure 3.
Molecular Dynamics Simulation of Cotranslational Folding of ADR1a
(A) The probability that the ADR1a domain is folded at 37°C is plotted as a function of L.
(B) Distance distribution of the folded (root-mean-square deviation [rmsd] < 3.5 Å from the cryo-EM ADR1a model, after alignment of the isolated domains; red) and unfolded (rmsd > 5.5 Å from the cryo-EM ADR1a model; blue) ADR1a domains in the exit tunnel. Distance distributions were calculated as a function of the distance between the last P atom in the tRNA of the cryo-EM structure and the Cα of His21 in ADR1a (2Å bins). Alignment of the cryo-EM and simulated ribosome structures was performed in advance. Top: Simulation run at 310 K. Bottom: Simulation run at 140 K. The arrow indicates the distance between the tRNA and His21 in the cryo-EM reconstruction.
(C) Snapshot of the folded structure of ADR1a (green) from the 140 K simulation that best overlaps the cryo-EM structure (red) in the exit tunnel at tether length L = 25. His21 is displayed in in its coarse-grained two-ball representation for the simulation model and in ball-and-stick representation for the cryo-EM structure.