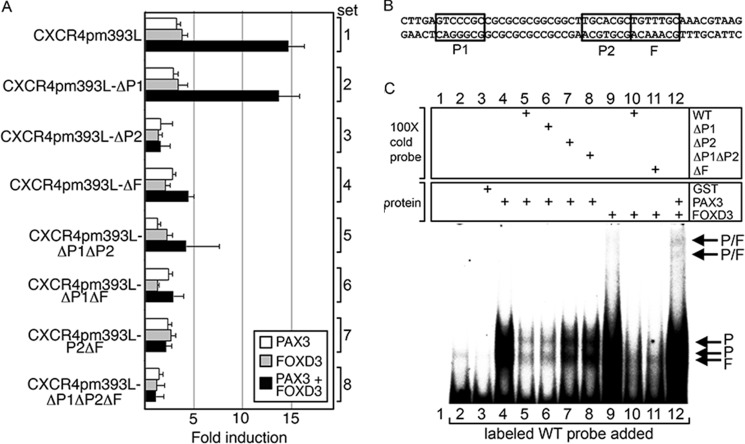
FIGURE 4.
The CXCR4 gene intronic enhancer region has specific PAX and FOX binding sites. A, mutations in putative PAX and FOX sites within the CXCR4 intronic enhancer alter the ability of PAX3 and FOXD3 to drive expression. pGL2-CXCR4pm393L or constructs with mutant P1, P2, and/or F sites were transfected into 293T cells with PAX3 (white), FOXD3 (gray), or both PAX3 and FOXD3 (black) expression constructs. Luciferase levels shown as fold-units over controls (levels without PAX3 or FOXD3 expression). B, sequence of the probe used in C with the PAX and FOX sites boxed. C, EMSA analysis for PAX3 and FOXD3 binding in vitro. Labeled probe encompassing the CXCR4 enhancer region as shown in B was incubated without any protein (lane 2), or with GST alone (lane 3) as negative controls. Lane 1 is without protein or probe (empty lane, negative control). The labeled probe was incubated with GST-PAX3 (lanes 4–8) or GST-FOXD3 (lane 9-11), or both (lane 12), and in the presence of ×100 cold probes with or without PAX or FOX sites mutated as indicated (lanes 5–8, 10, and 11), which may inhibit protein binding to the labeled probe.